













| General Information: |
|
| Name(s) found: |
RIP1 /
YEL024W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 215 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial respiratory chain complex III [IDA |
| Biological Process: |
mitochondrial electron transport, ubiquinol to cytochrome c
[IDA aerobic respiration [IMP |
| Molecular Function: |
ubiquinol-cytochrome-c reductase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLGIRSSVKT CFKPMSLTSK RLISQSLLAS KSTYRTPNFD DVLKENNDAD KGRSYAYFMV 60
61 GAMGLLSSAG AKSTVETFIS SMTATADVLA MAKVEVNLAA IPLGKNVVVK WQGKPVFIRH 120
121 RTPHEIQEAN SVDMSALKDP QTDADRVKDP QWLIMLGICT HLGCVPIGEA GDFGGWFCPC 180
181 HGSHYDISGR IRKGPAPLNL EIPAYEFDGD KVIVG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
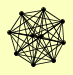
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
COB COR1 CYT1 QCR10 QCR2 QCR6 QCR7 QCR8 QCR9 RIP1 |
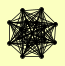
| View Details | Ho Y, et al. (2002) |
|
AHA1 AHP1 ARF1 GND1 GPG1 HEF3 HTB1 LHS1 MGE1 MGT1 RIP1 SEC27 SOF1 TMA19 UBR1 YHR033W |
| View Details | Qiu et al. (2008) |

|
AAC1 AAC3 ATP1 ATP14 ATP15 ATP16 ATP17 ATP18 ATP19 ATP2 ATP20 ATP3 ATP4 ATP5 ATP6 ATP7 ATP8 COB COR1 COX1 COX13 COX2 COX3 COX4 COX5A COX5B COX6 COX7 COX8 COX9 CYB2 CYC1 CYT1 INH1 MCR1 OLI1 POR1 QCR10 QCR2 QCR6 QCR7 QCR8 QCR9 RIP1 SDH1 SDH2 SDH3 SDH4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
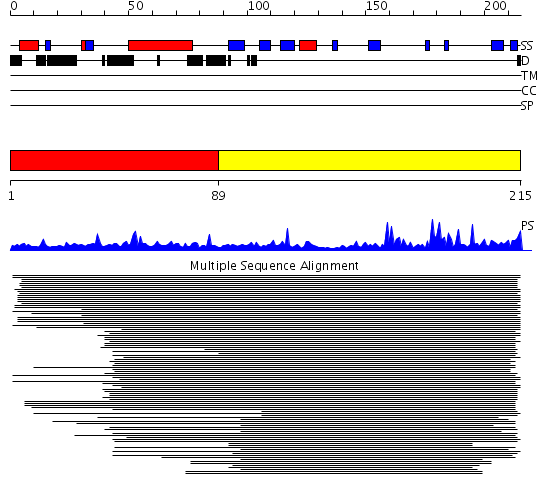
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 1149.69897 | ISP subunit of the mitochondrial cytochrome bc1-complex, watersoluble domain; Iron-sulfur subunit (ISP) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane region | |
| 2 | View Details | [89..215] | 1149.69897 | ISP subunit of the mitochondrial cytochrome bc1-complex, watersoluble domain; Iron-sulfur subunit (ISP) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane region |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)