













| General Information: |
|
| Name(s) found: |
COR1 /
YBL045C
[SGD]
|
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 457 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial respiratory chain complex III [IDA |
| Biological Process: |
aerobic respiration
[IMP |
| Molecular Function: |
ubiquinol-cytochrome-c reductase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRTVTSKTV SNQFKRSLAT AVATPKAEVT QLSNGIVVAT EHNPSAHTAS VGVVFGSGAA 60
61 NENPYNNGVS NLWKNIFLSK ENSAVAAKEG LALSSNISRD FQSYIVSSLP GSTDKSLDFL 120
121 NQSFIQQKAN LLSSSNFEAT KKSVLKQVQD FEENDHPNRV LEHLHSTAFQ NTPLSLPTRG 180
181 TLESLENLVV ADLESFANNH FLNSNAVVVG TGNIKHEDLV NSIESKNLSL QTGTKPVLKK 240
241 KAAFLGSEVR LRDDTLPKAW ISLAVEGEPV NSPNYFVAKL AAQIFGSYNA FEPASRLQGI 300
301 KLLDNIQEYQ LCDNFNHFSL SYKDSGLWGF STATRNVTMI DDLIHFTLKQ WNRLTISVTD 360
361 TEVERAKSLL KLQLGQLYES GNPVNDANLL GAEVLIKGSK LSLGEAFKKI DAITVKDVKA 420
421 WAGKRLWDQD IAIAGTGQIE GLLDYMRIRS DMSMMRW |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
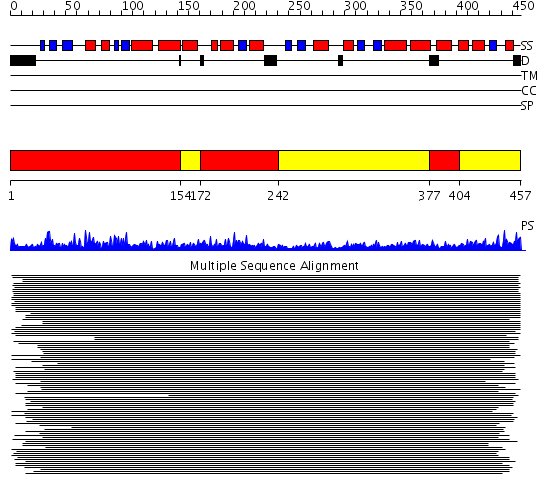
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] [172..241] [377..403] |
10090.69897 | Cytochrome bc1 core subunit 1 | |
| 2 | View Details | [154..171] [242..376] [404..457] |
10090.69897 | Cytochrome bc1 core subunit 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)