













| General Information: |
|
| Name(s) found: |
CPR6 /
YLR216C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 371 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
protein folding
[IDA |
| Molecular Function: |
peptidyl-prolyl cis-trans isomerase activity
[IDA unfolded protein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTRPKTFFDI SIGGKPQGRI VFELYNDIVP KTAENFLKLC EGNAGMAKTK PDVPLSYKGS 60
61 IFHRVIKDFM CQFGDFTNFN GTGGESIYDE KFEDENFTVK HDKPFLLSMA NAGPNTNGSQ 120
121 AFITCVPTPH LDGKHVVFGE VIQGKRIVRL IENQQCDQEN NKPLRDVKID DCGVLPDDYQ 180
181 VPENAEATPT DEYGDNYEDV LKQDEKVDLK NFDTVLKAIE TVKNIGTEQF KKQNYSVALE 240
241 KYVKCDKFLK EYFPEDLEKE QIEKINQLKV SIPLNIAICA LKLKDYKQVL VASSEVLYAE 300
301 AADEKAKAKA LYRRGLAYYH VNDTDMALND LEMATTFQPN DAAILKAIHN TKLKRKQQNE 360
361 KAKKSLSKMF S |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
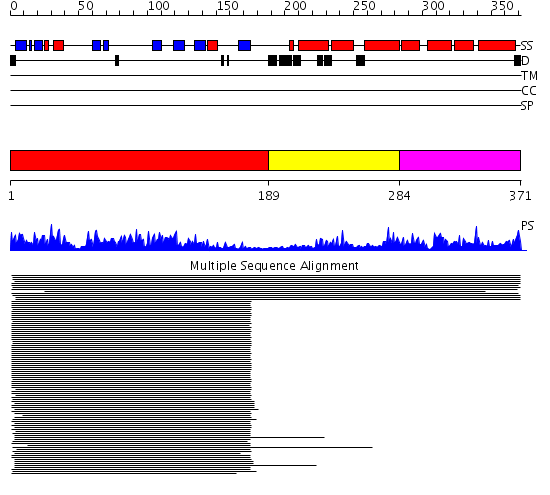
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..188] | 919.218487 | Cyclophilin 40; Cyclophilin 40 isomerase domain | |
| 2 | View Details | [189..283] | 919.218487 | Cyclophilin 40; Cyclophilin 40 isomerase domain | |
| 3 | View Details | [284..371] | 919.218487 | Cyclophilin 40; Cyclophilin 40 isomerase domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)