













| General Information: |
|
| Name(s) found: |
ADE5,7 /
YGL234W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 802 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
purine nucleotide biosynthetic process
[TAS]
'de novo' IMP biosynthetic process [TAS] purine base metabolic process [TAS |
| Molecular Function: |
phosphoribosylamine-glycine ligase activity
[TAS]
phosphoribosylformylglycinamidine cyclo-ligase activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNILVLGNG AREHVLVTKL AQSPTVGKIY VAPGNGGTAT MDPSRVINWD ITPDVANFAR 60
61 LQSMAVEHKI NLVVPGPELP LVNGITSVFH SVGIPVFGPS VKAAQLEASK AFSKRFMSKH 120
121 NIPTASYDVF TNPEEAISFL QAHTDKAFVI KADGIAAGKG VIIPSSIDES VQAIKDIMVT 180
181 KQFGEEAGKQ VVIEQFLEGD EISLLTIVDG YSHFNLPVAQ DHKRIFDGDK GLNTGGMGAY 240
241 APAPVATPSL LKTIDSQIVK PTIDGMRRDG MPFVGVLFTG MILVKDSKTN QLVPEVLEYN 300
301 VRFGDPETQA VLSLLDDQTD LAQVFLAAAE HRLDSVNIGI DDTRSAVTVV VAAGGYPESY 360
361 AKGDKITLDT DKLPPHTQIF QAGTKYDSAT DSLLTNGGRV LSVTSTAQDL RTAVDTVYEA 420
421 VKCVHFQNSY YRKDIAYRAF QNSESSKVAI TYADSGVSVD NGNNLVQTIK EMVRSTRRPG 480
481 ADSDIGGFGG LFDLAQAGFR QNEDTLLVGA TDGVGTKLII AQETGIHNTV GIDLVAMNVN 540
541 DLVVQGAEPL FFLDYFATGA LDIQVASDFV SGVANGCIQS GCALVGGETS EMPGMYPPGH 600
601 YDTNGTAVGA VLRQDILPKI NEMAAGDVLL GLASSGVHSN GFSLVRKIIQ HVALPWDAPC 660
661 PWDESKTLGE GILEPTKIYV KQLLPSIRQR LLLGLAHITG GGLVENIPRA IPDHLQARVD 720
721 MSTWEVPRVF KWFGQAGNVP HDDILRTFNM GVGMVLIVKR ENVKAVCDSL TEEGEIIWEL 780
781 GSLQERPKDA PGCVIENGTK LY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
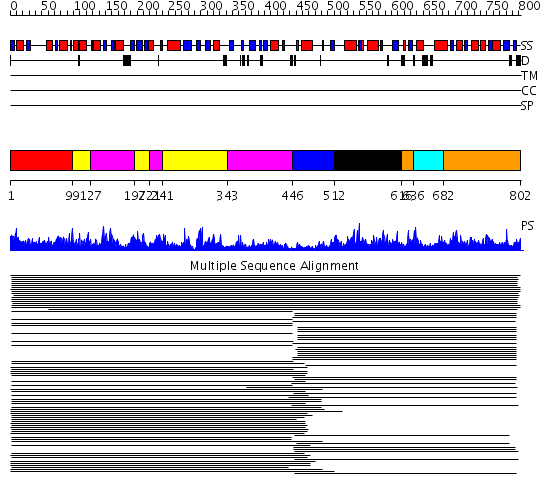
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 1021.0 | Glycinamide ribonucleotide synthetase (GAR-syn), C-domain; Glycinamide ribonucleotide synthetase (GAR-syn), N-domain; Glycinamide ribonucleotide synthetase (GAR-syn), domain 2 | |
| 2 | View Details | [99..126] [197..220] [241..342] |
1021.0 | Glycinamide ribonucleotide synthetase (GAR-syn), C-domain; Glycinamide ribonucleotide synthetase (GAR-syn), N-domain; Glycinamide ribonucleotide synthetase (GAR-syn), domain 2 | |
| 3 | View Details | [127..196] [221..240] [343..445] |
1021.0 | Glycinamide ribonucleotide synthetase (GAR-syn), C-domain; Glycinamide ribonucleotide synthetase (GAR-syn), N-domain; Glycinamide ribonucleotide synthetase (GAR-syn), domain 2 | |
| 4 | View Details | [446..511] | 7.0 | Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain; Aminoimidazole ribonucleotide synthetase (PurM) C-terminal domain | |
| 5 | View Details | [512..615] | 982.0 | Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain; Aminoimidazole ribonucleotide synthetase (PurM) C-terminal domain | |
| 6 | View Details | [616..635] [682..802] |
982.0 | Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain; Aminoimidazole ribonucleotide synthetase (PurM) C-terminal domain | |
| 7 | View Details | [636..681] | 982.0 | Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain; Aminoimidazole ribonucleotide synthetase (PurM) C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)