













| General Information: |
|
| Name(s) found: |
RSC2 /
YLR357W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 889 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RSC complex
[IDA |
| Biological Process: |
sister chromatid cohesion
[IMP ATP-dependent chromatin remodeling [IDA telomere maintenance [IMP double-strand break repair via nonhomologous end joining [IPI |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMPDDNSNSS TQNSSALYKD LRKEYESLFT LKEDSGLEIS PIFNVLPPKK DYPDYYAVIK 60
61 NPVSFNTLKK RIPHYTDAQQ FMNDVVQIPW NAKTYNTRDS GIYKYALVLE KYLKDTIYPN 120
121 LKEKYPQLVY PDLGPLPDEP GYEEFQQKLR EKAEEVARAN AARAESSSSM NSTEAARRLR 180
181 KTRTSVKRES EPGTDTNNDE DYEATDMDID NPKDADFPDL IRKPLININP YTRKPLRDNR 240
241 STTPSHSGTP QPLGPRHRQV SRTQVKRGRP PIIDLPYIQR MKNVMKVLKK EVLDSGIGLT 300
301 DLFERLPDRH RDANYYIMIA NPISLQDINK KVKTRRYKTF QEFQNDFNLM LTNFRISHRG 360
361 DPESIKISNI LEKTFTSLAR FELSKPDRSF IPEGELRYPL DEVIVNNISY HVGDWALLRN 420
421 QNDPQKPIVG QIFRLWKTPD GKQWLNACWY YRPEQTVHRV DRLFYKNEVM KTGQYRDHLV 480
481 SNLVGKCYVI HFTRYQRGNP DMKLEGPLFV CEFRYNESDK IFNKIRTWKA CLPEEIRDLD 540
541 EATIPVNGRK FFKYPSPIRH LLPANATPHD RVPEPTMGSP DAPPLVGAVY MRPKMQRDDL 600
601 GEYATSDDCP RYIIRPNDSP EEGQVDIETG TITTNTPTAN ALPKTGYSSS KLSSLRYNRS 660
661 SMSLENQNAI GQQQIPLSRV GSPGAGGPLT VQGLKQHQLQ RLQQQQHQYQ QQKRSQASRY 720
721 NIPTIIDDLT SQASRGNLGN IMIDAASSFV LPISITKNVD VLQRTDLHSQ TKRSGREEMF 780
781 PWKKTKGEIL WFRGPSVIVN ERIINSGDPH LSLPLNRWFT TNKKRKLEYE EVEETMEDVT 840
841 GKDKDDDGLE PDVENEKESL PGPFVLGLRP SAKFTAHRLS MLRPPSSSS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP7 ARP9 HTL1 ISW1 ITC1 MOT1 NPL6 RSC1 RSC2 RSC3 RSC30 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 VPS1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ARP7 ARP9 RSC1 RSC2 RSC4 RSC6 RSC8 SAS5 SFH1 STH1 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP7 ARP9 ATG16 CSH1 GRS2 HTL1 MGR1 NFI1 NPL6 PCL9 RIM15 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 URC2 YIL177C |
| View Details | Gavin AC, et al. (2006) |
|
ARP7 NPL6 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 SFH1 STH1 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ADE5,7 ADH1 ARP4 ARP8 BEM1 BOI2 CDC24 ESC8 GCN1 HHF1, HHF2 IES1 IES5 INO80 IOC3 ISW1 ISW2 ITC1 MOT1 NHP10 NPL6 PDC1 REB1 RFA1 RFX1 RNR2 RPC34 RPC40 RPO31 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RVB2 SFH1 SIN3 SPT15 STH1 STO1 TAF1 TFC7 TOP1 VPS1 YBR090C-A, NHP6B |
| View Details | Qiu et al. (2008) |
|
ARP7 ARP9 NPL6 RSC1 RSC2 RSC4 RSC58 RSC6 RSC8 TAF14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
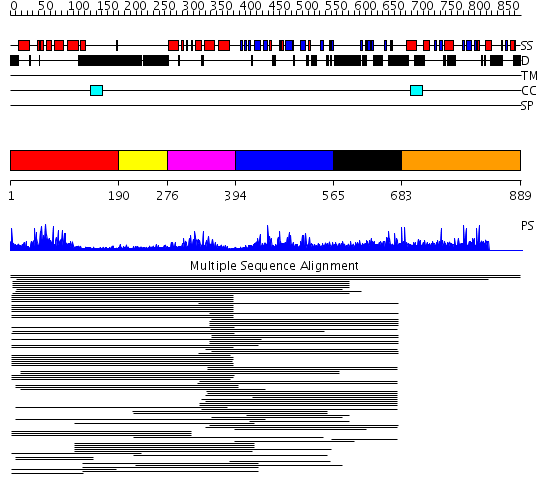
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | 60.67837 | GCN5 | |
| 2 | View Details | [190..275] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [276..393] | 23.920819 | Bromodomain No confident structure predictions are available. | |
| 4 | View Details | [394..564] | 40.721246 | BAH domain No confident structure predictions are available. | |
| 5 | View Details | [565..682] | 3.04098 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [683..889] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)