













| General Information: |
|
| Name(s) found: |
INO80 /
YGL150C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1489 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Ino80 complex
[IDA |
| Biological Process: |
DNA repair
[IDA chromatin remodeling [IDA transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
ATPase activity
[IDA ATP-dependent 5'-3' DNA helicase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLAVLLNKE DKDISDFSKT TAGKSAKKNS RERVADVAPT RVLDKKQAYL SQLNSEFNRI 60
61 KRRDSIEQLY QDWKFINLQE FELISEWNQQ SKDWQFDNTN DSQDLHFKKL YRDMSMINKE 120
121 WAEYQSFKNA NLSDIINEKD ADEDEEDDED ELEDGEEDME EDEASTGRHT NGKSMRGNGI 180
181 QKSRKKDAAA AAAIGKAIKD DQTHADTVVT VNGDENEDGN NGEDEDNDND NENNNDNDND 240
241 NENENDNDSD NDDEEENGEE DEEEEEIEDL DEEDFAAFEE QDDNDDEDFN PDVEKRRKRS 300
301 SSSSSSTKLS MNSLSLITSK KINKNITINS DRPKIVRELI KMCNKNKHQK IKKRRFTNCI 360
361 VTDYNPIDSK LNIKITLKQY HVKRLKKLIN DAKREREREE ALKNNVGLDG NDLDNDEDGS 420
421 ESHKRRKLNN NTANGADDAN KRKFNTRHGL PTYGMKMNAK EARAIQRHYD NTYTTIWKDM 480
481 ARKDSTKMSR LVQQIQSIRS TNFRKTSSLC AREAKKWQSK NFKQIKDFQT RARRGIREMS 540
541 NFWKKNEREE RDLKKKIEKE AMEQAKKEEE EKESKRQAKK LNFLLTQTEL YSHFIGRKIK 600
601 TNELEGNNVS SNDSESQKNI DISALAPNKN DFHAIDFDNE NDEQLRLRAA ENASNALAET 660
661 RAKAKQFDDH ANAHEEEEEE DELNFQNPTS LGEITIEQPK ILACTLKEYQ LKGLNWLANL 720
721 YDQGINGILA DEMGLGKTVQ SISVLAHLAE NHNIWGPFLV VTPASTLHNW VNEISKFLPQ 780
781 FKILPYWGNA NDRKVLRKFW DRKNLRYNKN APFHVMVTSY QMVVTDANYL QKMKWQYMIL 840
841 DEAQAIKSSQ SSRWKNLLSF HCRNRLLLTG TPIQNSMQEL WALLHFIMPS LFDSHDEFNE 900
901 WFSKDIESHA EANTKLNQQQ LRRLHMILKP FMLRRVKKNV QSELGDKIEI DVLCDLTQRQ 960
961 AKLYQVLKSQ ISTNYDAIEN AATNDSTSNS ASNSGSDQNL INAVMQFRKV CNHPDLFERA1020
1021 DVDSPFSFTT FGKTTSMLTA SVANNNSSVI SNSNMNLSSM SSNNISNGKF TDLIYSSRNP1080
1081 IKYSLPRLIY EDLILPNYNN DVDIANKLKN VKFNIFNPST NYELCLFLSK LTGEPSLNEF1140
1141 FRVSTTPLLK RVIERTNGPK NTDSLSFKTI TQELLEVTRN APSEGVMASL LNVEKHAYER1200
1201 EYLNCIQRGY HPNVSAPPVT IEVLGSSHVT NSINNELFDP LISQALSDIP AITQYNMHVK1260
1261 KGIPVEDFPK TGLFPEPLNK NFSSNISMPS MDRFITESAK LRKLDELLVK LKSEGHRVLI1320
1321 YFQMTKMMDL MEEYLTYRQY NHIRLDGSSK LEDRRDLVHD WQTNPEIFVF LLSTRAGGLG1380
1381 INLTAADTVI FYDSDWNPTI DSQAMDRAHR LGQTRQVTVY RLLVRGTIEE RMRDRAKQKE1440
1441 QVQQVVMEGK TQEKNIKTIE VGENDSEVTR EGSKSISQDG IKEAASALA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ACT1 ARP4 ARP5 ARP8 IES1 IES3 IES5 INO80 NHP10 RVB1 RVB2 TAF14 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
| View Details | Gavin AC, et al. (2006) |
|
ARP5 ARP8 IES1 IES3 IES5 INO80 NHP10 |
| View Details | Gavin AC, et al. (2002) |

|
ACT1 ADH1 ARP4 ARP8 ESC8 IES1 IES5 INO80 IOC3 ISW1 ISW2 ITC1 MOT1 NHP10 NPL6 RPC40 RSC2 RSC4 RSC58 RSC6 RSC8 RVB2 SIN3 VPS1 |
| View Details | Ho Y, et al. (2002) |
|
ADH2 FKH2 HTB2 INO80 SIN3 |
| View Details | Qiu et al. (2008) |
|
ARP4 ARP5 INO80 RVB1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
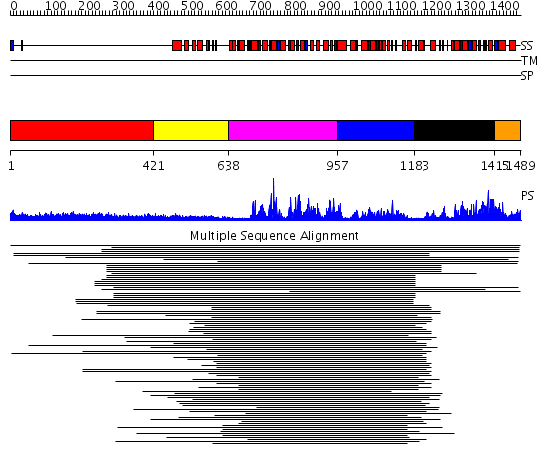
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..420] | 4.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [421..637] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [638..956] | 14.8 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [957..1182] | 14.8 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 5 | View Details | [1183..1414] | 2.39794 | Putative DEAD box RNA helicase | |
| 6 | View Details | [1415..1489] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
|||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)