













| General Information: |
|
| Name(s) found: |
EPL1 /
YFL024C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 832 amino acids |
Gene Ontology: |
|
| Cellular Component: |
NuA4 histone acetyltransferase complex
[IPI Piccolo NuA4 histone acetyltransferase complex [IDA histone acetyltransferase complex [IDA |
| Biological Process: |
DNA repair
[IDA histone acetylation [IDA regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
histone acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTPSNAIEI NDGSHKSGRS TRRSGSRSAH DDGLDSFSKG DSGAGASAGS SNSRFRHRKI 60
61 SVKQHLKIYL PNDLKHLDKD ELQQREVVEI ETGVEKNEEK EVHLHRILQM GSGHTKHKDY 120
121 IPTPDASMTW NEYDKFYTGS FQETTSYIKF SATVEDCCGT NYNMDERDET FLNEQVNKGS 180
181 SDILTEDEFE ILCSSFEHAI HERQPFLSMD PESILSFEEL KPTLIKSDMA DFNLRNQLNH 240
241 EINSHKTHFI TQFDPVSQMN TRPLIQLIEK FGSKIYDYWR ERKIEVNGYE IFPQLKFERP 300
301 GEKEEIDPYV CFRRREVRHP RKTRRIDILN SQRLRALHQE LKNAKDLALL VAKRENVSLN 360
361 WINDELKIFD QRVKIKNLKR SLNISGEDDD LINHKRKRPT IVTVEQREAE LRKAELKRAA 420
421 AAAAAAKAKN NKRNNQLEDK SSRLTKQQQQ QLLQQQQQQQ QNALKTENGK QLANASSSST 480
481 SQPITSHVYV KLPSSKIPDI VLEDVDALLN SKEKNARKFV QEKMEKRKIE DADVFFNLTD 540
541 DPFNPVFDMS LPKNFSTSNV PFASIASSKF QIDRSFYSSH LPEYLKGISD DIRIYDSNGR 600
601 SRNKDNYNLD TKRIKKTELY DPFQENLEIH SREYPIKFRK RVGRSNIKYV DRMPNFTTSS 660
661 TKSACSLMDF VDFDSIEKEQ YSREGSNDTD SINVYDSKYD EFVRLYDKWK YDSPQNEYGI 720
721 KFSDEPARLN QISNDTQVIR FGTMLGTKSY EQLREATIKY RRDYITRLKQ KHIQHLQQQQ 780
781 QQQQQQQQQA QQQKQKSQNN NSNSSNSLKK LNDSLINSEA KQNSSITQKN SS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP4 EAF1 EAF3 EAF5 EAF6 EAF7 EPL1 ESA1 SWC4 TRA1 YAF9 YNG2 |
| View Details | Hazbun TR, et al. (2003) |
|
ARP4 EAF1 EAF5 EPL1 ESA1 RVB1 RVB2 SWC4 SWR1 TRA1 YAF9 YNG2 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
| View Details | Gavin AC, et al. (2006) |

|
EPL1 ESA1 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ADH1 ARP4 EAF1 EAF3 EAF5 EPL1 ESA1 HHF1, HHF2 TRA1 YAF9 YAP1 |
| View Details | Ho Y, et al. (2002) |
|
ARP4 ATE1 CDC33 CKA2 EAF1 EPL1 ESA1 HRR25 POR1 SAP185 SIT4 TAF14 TAF4 TIP41 TRA1 TUF1 YRA1 |
| View Details | Qiu et al. (2008) |
|
AHC1 ARP4 EAF3 EPL1 ESA1 GCN5 SAS3 SGF73 SPT7 SWC4 TAF1 TAF10 TAF12 TAF6 TRA1 YNG1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
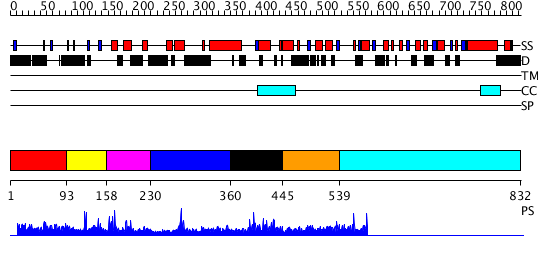
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [93..157] | 1.020991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [158..229] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [230..359] | 12.154902 | TAFII250 double bromodomain module | |
| 5 | View Details | [360..444] | 12.154902 | TAFII250 double bromodomain module | |
| 6 | View Details | [445..538] | 3.17299 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [539..832] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)