













| General Information: |
|
| Name(s) found: |
YNG2 /
YHR090C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 282 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA NuA4 histone acetyltransferase complex [IPI Piccolo NuA4 histone acetyltransferase complex [IDA histone acetyltransferase complex [IDA |
| Biological Process: |
chromatin modification
[IMP DNA repair [IDA histone acetylation [IDA |
| Molecular Function: |
enzyme activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPSLVLEQT IQDVSNLPSE FRYLLEEIGS NDLKLIEEKK KYEQKESQIH KFIRQQGSIP 60
61 KHPQEDGLDK EIKESLLKCQ SLQREKCVLA NTALFLIARH LNKLEKNIAL LEEDGVLAPV 120
121 EEDGDMDSAA EASRESSVVS NSSVKKRRAA SSSGSVPPTL KKKKTSRTSK LQNEIDVSSR 180
181 EKSVTPVSPS IEKKIARTKE FKNSRNGKGQ NGSPENEEED KTLYCFCQRV SFGEMVACDG 240
241 PNCKYEWFHY DCVNLKEPPK GTWYCPECKI EMEKNKLKRK RN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP4 BDF1 EAF1 EAF3 EAF5 EAF6 EAF7 EPL1 ESA1 SWC4 YAF9 YNG2 |
| View Details | Hazbun TR, et al. (2003) |
|
ARP4 EAF1 EAF5 EPL1 ESA1 RVB1 RVB2 SWC4 SWR1 TRA1 YAF9 YNG2 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
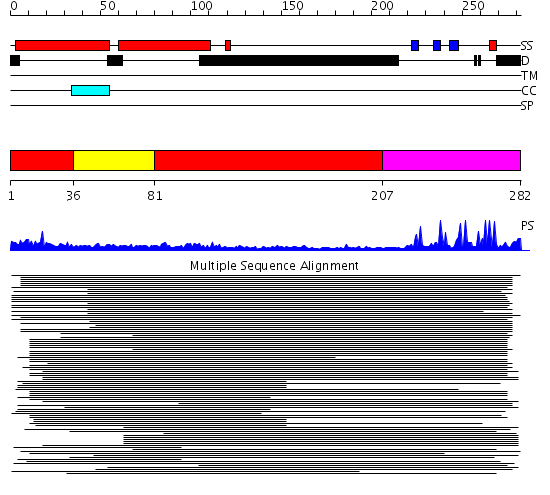
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..35] [81..206] |
7.77 | Variant surface glycoprotein (N-terminal domain) | |
| 2 | View Details | [36..80] | 7.77 | Variant surface glycoprotein (N-terminal domain) | |
| 3 | View Details | [207..282] | 5.0 | Williams-Beuren syndrome transcription factor, WSTF |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)