













| General Information: |
|
| Name(s) found: |
IES3 /
YLR052W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 250 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Ino80 complex [IPI |
| Biological Process: |
chromatin remodeling
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFEDLLATN KQVQFAHAAT QHYKSVKTPD FLEKDPHHKK FHNADGLNQQ GSSTPSTATD 60
61 ANAASTASTH TNTTTFKRHI VAVDDISKMN YEMIKNSPGN VITNANQDEI DISTLKTRLY 120
121 KDNLYAMNDN FLQAVNDQIV TLNAAEQDQE TEDPDLSDDE KIDILTKIQE NLLEEYQKLS 180
181 QKERKWFILK ELLLDANVEL DLFSNRGRKA SHPIAFGAVA IPTNVNANSL AFNRTKRRKI 240
241 NKNGLLENIL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
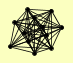
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP4 ARP5 ARP8 IES1 IES2 IES3 IES5 INO80 NHP10 RVB1 RVB2 TAF14 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
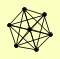
| View Details | Gavin AC, et al. (2006) |
|
ARP5 ARP8 IES1 IES3 IES5 INO80 NHP10 |
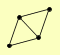
| View Details | Qiu et al. (2008) |
|
ARP5 IES3 RVB1 TAF14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
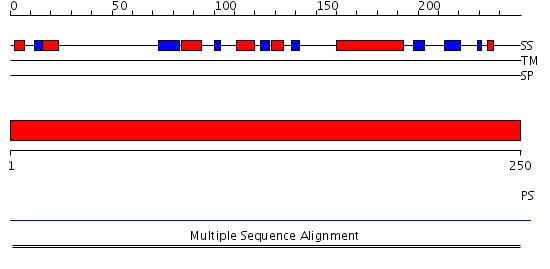
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..250] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)