













| General Information: |
|
| Name(s) found: |
EAF5 /
YEL018W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 279 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA NuA4 histone acetyltransferase complex [IPI |
| Biological Process: |
DNA repair
[IDA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKEVSELVV LQLIHTLISN KNEELVRNGG GINMIGNNLR ISLVKLTNEI QNNLLINELT 60
61 NLRRQSNVAN GNRKLGINDI LTIVKNLFPE YRTTLNDGQL SLHGLEMHDI EKLLDEKYDR 120
121 FKKTQVEQIR MMEDEILKNG IKTGASQLQP HANAGKSGSA GTSATITTTT PHMAHSMDPK 180
181 REKLLKLYRD TVLNKLESKT GNFQKLFKSP DGSIIKNEIN YEDIKNETPG SVHELQLILQ 240
241 KSITDGVMRK VIGTDDWKLA RQVQFELDDT VQFMRRALE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP4 EAF1 EAF3 EAF5 EAF6 EAF7 EPL1 ESA1 SRP1 SWC4 TRA1 YAF9 YNG2 |
| View Details | Hazbun TR, et al. (2003) |
|
ARP4 EAF1 EAF5 EPL1 ESA1 RVB1 RVB2 SWC4 SWR1 TRA1 YAF9 YNG2 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ADH1 ARP4 EAF1 EAF3 EAF5 EPL1 ESA1 HHF1, HHF2 TRA1 YAF9 YAP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
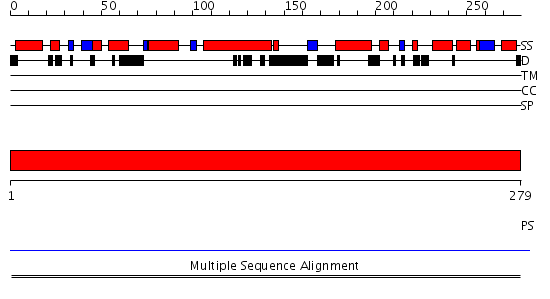
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..279] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)