













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGIMVGMGQK DSYVGDEAQS 60
61 KRGILTLRYP IEHGIVTNWD DMEKIWHHTF YNELRVAPEE HPVLLTEAPM NPKSNREKMT 120
121 QIMFETFNVP AFYVSIQAVL SLYSSGRTTG IVLDSGDGVT HVVPIYAGFS LPHAILRIDL 180
181 AGRDLTDYLM KILSERGYSF STTAEREIVR DIKEKLCYVA LDFEQEMQTA AQSSSIEKSY 240
241 ELPDGQVITI GNERFRAPEA LFHPSVLGLE SAGIDQTTYN SIMKCDVDVR KELYGNIVMS 300
301 GGTTMFPGIA ERMQKEITAL APSSMKVKII APPERKYSVW IGGSILASLT TFQQMWISKQ 360
361 EYDESGPSIV HHKCF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
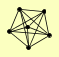
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ACT1 ARP8 IES1 INO80 ISW1 NHP10 |
| View Details | Gavin AC, et al. (2006) |

|
ACT1 MGS1 SAC6 |
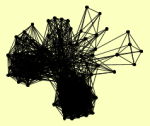
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ADH1 AIM31 APC1 APC2 ARP4 ARP8 ARX1 CDC16 CDC23 CDC27 CFT1 CFT2 CKA1 CLP1 CYR1 EAF1 EAF3 EAF5 ENO2 EPL1 ERG13 ESA1 ESC8 FIP1 FPR4 GCN1 GLC7 HHF1, HHF2 HSC82 HTA1 IES1 IES5 INO80 IOC3 ISW1 ISW2 ITC1 KAP123 LYS12 MLC1 MLC2 MOT1 MPE1 MYO1 MYO2 MYO4 NAN1 NCS2 NHP10 NIP1 NPL6 PAP1 PCF11 PFK1 PFS2 PHO81 POL5 PTA1 PTI1 PWP1 REF2 RNA14 RNA15 RPC40 RRN3 RSA3 RSC2 RSC4 RSC58 RSC6 RSC8 RVB2 SAM1 SEC13 SEC31 SHE2 SHE3 SIN3 SMC1 SRV2 SSA3 SSU72 SYC1 TAF6 TIF4632 TRA1 TYE7 TYW1 VID24 VMR1 VPS1 VPS53 YAF9 YAP1 YCL046W YEF3 YSH1 YTH1 |
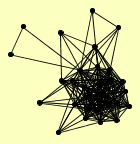
| View Details | Qiu et al. (2008) |
|
ABP1 ACT1 ARC40 ARP2 ARP3 ARP4 BEM1 BNI1 CAP1 CAP2 CDC11 CDC12 COF1 DLD2 LAS17 MYO1 MYO2 MYO5 OYE2 PAN1 PFY1 RVS167 SAC6 SCD5 SLA2 SRV2 TPM1 TPM2 TRA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
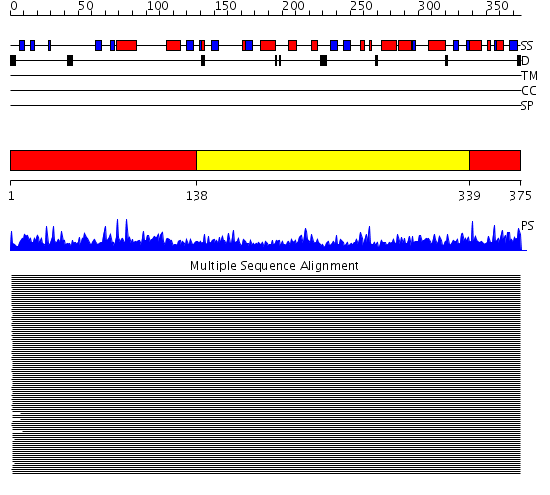
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] [339..375] |
11000.0 | Actin | |
| 2 | View Details | [138..338] | 11000.0 | Actin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)