













| General Information: |
|
| Name(s) found: |
TYW1 /
YPL207W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGFRVAGAL VVGALTAAYL YFGGRFSIAL VIIVGYGIYC NEASGGSQDS QEKLDLNKQQ 60
61 KKPCCSDKKI ADGGKKTGGC CSDKKNGGGK GGGCCSSKGG KKGGCCSSKG GKKGGCCSSK 120
121 KNIGDNENTA TEVEKAVNYP VTVDFTEVFR KPTKKRSSTP KVFSKNSSSN SRVGKKLSVS 180
181 KKIGPDGLIK SALTISNETL LSSQIYVLYS SLQGAASKAA KSVYDKLKEL DELTNEPKLL 240
241 NLDDLSDFDD YFINVPVENA LYVLVLPSYD IDCPLDYFLQ TLEENANDFR VDSFPLRKLV 300
301 GYTVLGLGDS ESWPEKFCYQ AKRADHWISR LGGRRIFPLG KVCMKTGGSA KIDEWTSLLA 360
361 ETLKDDEPII YEYDENADSE EDEEEGNGSD ELGDVEDIGG KGSNGKFSGA DEIKQMVAKD 420
421 SPTYKNLTKQ GYKVIGSHSG VKICRWTKNE LRGKGSCYKK SLFNIASSRC MELTPSLACS 480
481 SKCVFCWRHG TNPVSKNWRW EVDEPEYILE NALKGHYSMI KQMRGVPGVI AERFAKAFEV 540
541 RHCALSLVGE PILYPHINKF IQLLHQKGIT SFLVCNAQHP EALRNIVKVT QLYVSIDAPT 600
601 KTELKKVDRP LYKDFWERMV ECLEILKTVQ NHQRTVFRLT LVKGFNMGDV SAYADLVQRG 660
661 LPGFIEVKGA TFSGSSDGNG NPLTMQNIPF YEECVKFVKA FTTELQRRGL HYDLAAEHAH 720
721 SNCLLIADTK FKINGEWHTH IDFDKFFVLL NSGKDFTYMD YLEKTPEWAL FGNGGFAPGN 780
781 TRVYRKDKKK QNKENQETTT RETPLPPIPA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CSR1 TYW1 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ENO2 FPR4 NAN1 POL5 PWP1 SMC1 TYW1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
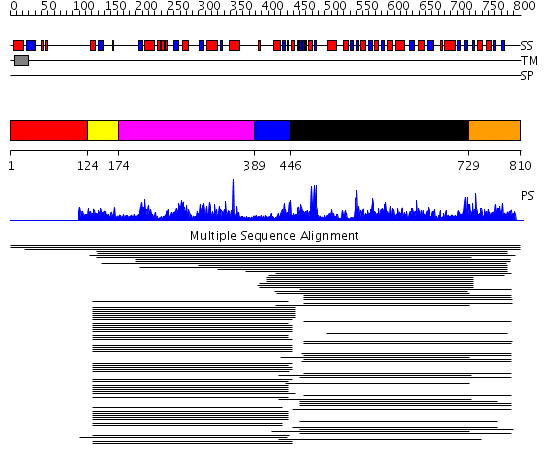
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [124..173] | 12.522879 | Nitric oxide (NO) synthase oxygenase domain | |
| 3 | View Details | [174..388] | 67.39794 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain | |
| 4 | View Details | [389..445] | 67.39794 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain | |
| 5 | View Details | [446..728] | 226.736997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [729..810] | 2.247995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 |
|
|||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|