













| General Information: |
|
| Name(s) found: |
CSR1 /
YLR380W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 408 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA microsome [IDA cytosol [IDA |
| Biological Process: |
cellular cell wall organization
[IGI phospholipid transport [IDA ER-associated protein catabolic process [IMP |
| Molecular Function: |
phosphatidylinositol transporter activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFDRQLTED QEVVLKQIWT HLFHLWQVPV DGTHIFPNNS LHSSSTPAKK KKSSWFSKLQ 60
61 SSDHTQDSSE AAEAAHLYEK GKIHKALANL DPQTTKKQFW HDIKNETPDA TILKFIRARK 120
121 WNADKTIAML GHDLYWRKDT INKIINGGER AVYENNETGV IKNLELQKAT IQGYDNDMRP 180
181 VILVRPRLHH SSDQTEQELE KFSLLVIEQS KLFFKENYPA STTILFDLNG FSMSNMDYAP 240
241 VKFLITCFEA HYPESLGHLL IHKAPWIFNP IWNIIKNWLD PVVASKIVFT KNIDELHKFI 300
301 QPQYIPRYLG GENDNDLDHY TPPDGSLDVH LKDTETRAMI EKEREELVEQ FLTVTAQWIE 360
361 HQPLNDPAYI QLQEKRVQLS TALCENYSKL DPYIRSRSVY DYNGSLKV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CHA1 CSR1 KAP123 RPL8B RPS0A SEC65 |
| View Details | Krogan NJ, et al. (2006) |

|
CSR1 TYW1 |
| View Details | Qiu et al. (2008) |

|
CSR1 FAT1 SFH5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
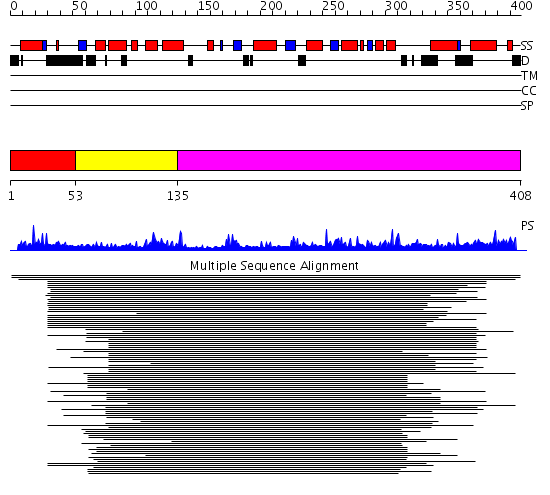
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..52] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [53..134] | 127.9897 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p | |
| 3 | View Details | [135..408] | 127.9897 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)