













| General Information: |
|
| Name(s) found: |
CHA1 /
YCL064C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 360 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial nucleoid [IDA |
| Biological Process: |
threonine catabolic process
[IMP serine family amino acid catabolic process [IMP |
| Molecular Function: |
L-serine ammonia-lyase activity
[IMP L-threonine ammonia-lyase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIVYNKTPL LRQFFPGKAS AQFFLKYECL QPSGSFKSRG IGNLIMKSAI RIQKDGKRSP 60
61 QVFASSGGNA GFAAATACQR LSLPCTVVVP TATKKRMVDK IRNTGAQVIV SGAYWKEADT 120
121 FLKTNVMNKI DSQVIEPIYV HPFDNPDIWE GHSSMIDEIV QDLKSQHISV NKVKGIVCSV 180
181 GGGGLYNGII QGLERYGLAD RIPIVGVETN GCHVFNTSLK IGQPVQFKKI TSIATSLGTA 240
241 VISNQTFEYA RKYNTRSVVI EDKDVIETCL KYTHQFNMVI EPACGAALHL GYNTKILENA 300
301 LGSKLAADDI VIIIACGGSS NTIKDLEEAL DSMRKKDTPV IEVADNFIFP EKNIVNLKSA 360
361 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CHA1 CSR1 |
| View Details | Krogan NJ, et al. (2006) |

|
CHA1 SDA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
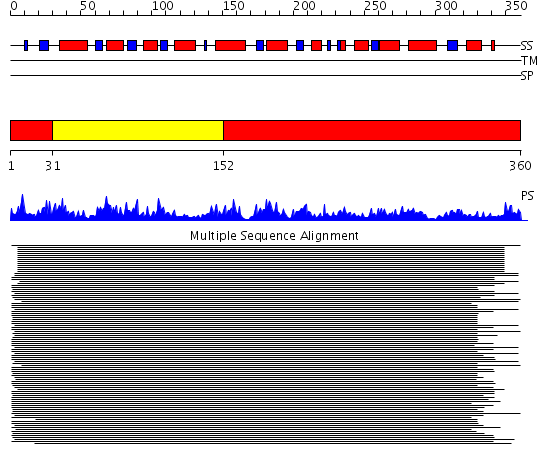
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..30] [152..360] |
201.502279 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain | |
| 2 | View Details | [31..151] | 201.502279 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)