













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSQPVDVDN IIDRLLEVRG SKPGQQVDLE ENEIRYLCSK ARSIFIKQPI LLELEAPIKI 60
61 CGDIHGQYYD LLRLFEYGGF PPESNYLFLG DYVDRGKQSL ETICLLLAYK IKYPENFFIL 120
121 RGNHECASIN RIYGFYDECK RRYNIKLWKT FTDCFNCLPI AAIIDEKIFC MHGGLSPDLN 180
181 SMEQIRRVMR PTDIPDVGLL CDLLWSDPDK DIVGWSENDR GVSFTFGPDV VNRFLQKQDM 240
241 ELICRAHQVV EDGYEFFSKR QLVTLFSAPN YCGEFDNAGA MMSVDESLLC SFQILKPAQK 300
301 SLPRQAGGRK KK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
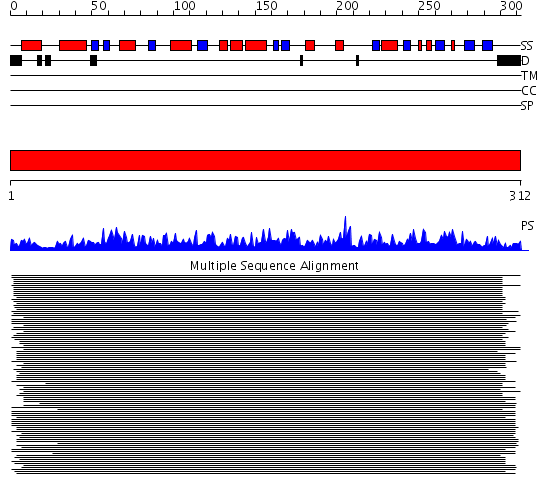
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..312] | 1734.0 | Protein phosphatase-1 (PP-1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)