













| General Information: |
|
| Name(s) found: |
EDE1 /
YBL047C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1381 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud tip
[IDA cellular bud neck [IDA actin cortical patch [IDA |
| Biological Process: |
endocytosis
[IMP |
| Molecular Function: |
ubiquitin binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASITFRTPL SSQEQAFYNQ KFHQLDTEDL GVVTGEAVRP LFASSGLPGQ LLSQVWATVD 60
61 IDNKGFLNLN EFSAALRMIA QLQNAPNQPI SAALYESTPT QLASFSINQN PAPMQSGSAT 120
121 GNTNNTDIPA LSSNDIAKFS QLFDRTAKGA QTVAGDKAKD IFLKARLPNQ TLGEIWALCD 180
181 RDASGVLDKS EFIMAMYLIQ LCMSHHPSMN TPPAVLPTQL WDSIRLEPVV VNQPNRTTPL 240
241 SANSTGVSSL TRHSTISRLS TGAFSNAASD WSLSFEKKQQ FDAIFDSLDK QHAGSLSSAV 300
301 LVPFFLSSRL NQETLATIWD LADIHNNAEF TKLEFAIAMF LIQKKNAGVE LPDVIPNELL 360
361 QSPALGLYPP NPLPQQQSAP QIAIPSRASK PSLQDMPHQV SAPAVNTQPT VPQVLPQNSN 420
421 NGSLNDLLAL NPSFSSPSPT KAQTVVQNNT NNSFSYDNNN GQATLQQQQP QQPPPLTHSS 480
481 SGLKKFTPTS NFGQSIIKEE PEEQEQLRES SDTFSAQPPP VPKHASSPVK RTASTTLPQV 540
541 PNFSVFSMPA GAATSAATGA AVGAAVGAAA LGASAFSRSS NNAFKNQDLF ADGEASAQLS 600
601 NATTEMANLS NQVNSLSKQA SITNDKKSRA TQELKRVTEM KNSIQIKLNN LRSTHDQNVK 660
661 QTEQLEAQVL QVNKENETLA QQLAVSEANY HAAESKLNEL TTDLQESQTK NAELKEQITN 720
721 LNSMTASLQS QLNEKQQQVK QERSMVDVNS KQLELNQVTV ANLQKEIDGL GEKISVYLTK 780
781 QKELNDYQKT VEEQHAQLQA KYQDLSNKDT DLTDREKQLE ERNRQIEEQE NLYHQHVSKL 840
841 QEMFDDLSQR KASFEKADQE LKERNIEYAN NVRELSERQM NLAMGQLPED AKDIIAKSAS 900
901 NTDTTTKEAT SRGNVHEDTV SKFVETTVEN SNLNVNRVKD DEEKTERTES DVFDRDVPTL 960
961 GSQSDSENAN TNNGTQSGNE TANPNLTETL SDRFDGDLNE YGIPRSQSLT SSVANNAPQS1020
1021 VRDDVELPET LEERDTINNT ANRDNTGNLS HIPGEWEATP ATASTDVLSN ETTEVIEDGS1080
1081 TTKRANSNED GESVSSIQES PKISAQPKAK TINEEFPPIQ ELHIDESDSS SSDDDEFEDT1140
1141 REIPSATVKT LQTPYNAQPT SSLEIHTEQV IKYPAPGTSP SHNEGNSKKA STNSILPVKD1200
1201 EFDDEFAGLE QAAVEEDNGA DSESEFENVA NAGSMEQFET IDHKDLDDEL QMNAFTGTLT1260
1261 SSSNPTIPKP QVQQQSTSDP AQVSNDEWDE IFAGFGNSKA EPTKVATPSI PQQPIPLKND1320
1321 PIVDASLSKG PIVNRGVATT PKSLAVEELS GMGFTEEEAH NALEKCNWDL EAATNFLLDS1380
1381 A |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
EDE1 SYP1 |
| View Details | Krogan NJ, et al. (2006) |

|
EDE1 SYP1 |
| View Details | Gavin AC, et al. (2006) |
|
EDE1 SYP1 |
| View Details | Gavin AC, et al. (2002) |
|
EDE1 SLA2 SYP1 YDR348C |
| View Details | Ho Y, et al. (2002) |

|
ABP1 ACO1 AIM41 ASF1 ATP3 ATP4 BEM2 BUD14 CAR2 CDC13 CDC25 CKA2 CMD1 CMK2 CMP2 COF1 COP1 COR1 CPR1 CRZ1 CYS4 DCP1 DCP2 DNM1 DUF1 EDC3 EDE1 EGD2 ENP1 FOL2 FPR3 GAC1 GAS1 GCN3 GLC7 GPH1 HCH1 HHF1, HHF2 HRR25 HSP104 HTA1 HTA2 HTB1 HTB2 HXT7 HYP2 ILS1 IPP1 KAP95 KNS1 LHS1 LOC1 LSP1 LTV1 MAM33 MDH1 MDS3 MGE1 MYO2 MYO4 MYO5 NIP1 NOB1 NSA1 NUG1 OYE2 PEX19 PGM2 PIL1 PIN4 POB3 POR1 PSH1 PST2 PTC2 PTC4 PUF3 RAD53 RIO2 RPC19 RPG1 RPM2 SAP185 SAP190 SAS10 SEC2 SEC23 SES1 SFB3 SGM1 SHE3 SIT4 SOD1 SPT16 SRP1 SRV2 SWI4 SYP1 TBF1 TGL1 TIF34 TIF35 TMA19 TSR1 UBA1 UBI4 UTP22 VAS1 VMA4 VPS13 YBR225W YER138C YNK1 YTA7 |
| View Details | Qiu et al. (2008) |
|
ARP2 EDE1 ENT1 ENT3 PAN1 RVS167 SAC6 SCD5 SLA1 SLA2 VRP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
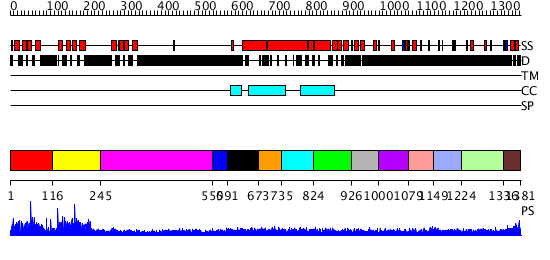
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 83.124002 | Eps15 | |
| 2 | View Details | [116..244] | 161.39794 | STRUCTURE OF THE SECOND EPS15 HOMOLOGY DOMAIN OF HUMAN EPS15, NMR, 20 STRUCTURES | |
| 3 | View Details | [245..549] | 66.07631 | Heavy meromyosin subfragment | |
| 4 | View Details | [550..590] | 35.67837 | Tropomyosin | |
| 5 | View Details | [591..672] | 35.67837 | Tropomyosin | |
| 6 | View Details | [673..734] | 35.67837 | Tropomyosin | |
| 7 | View Details | [735..823] | 35.67837 | Tropomyosin | |
| 8 | View Details | [824..925] | N/A | Confident ab initio structure predictions are available. | |
| 9 | View Details | [926..999] | 10.67 | No description for 1l8mA was found. | |
| 10 | View Details | [1000..1078] | 10.67 | No description for 1l8mA was found. | |
| 11 | View Details | [1079..1148] | 10.67 | No description for 1l8mA was found. | |
| 12 | View Details | [1149..1223] | 10.67 | No description for 1l8mA was found. | |
| 13 | View Details | [1224..1335] | 14.51 | No description for 1ldvA was found. | |
| 14 | View Details | [1336..1381] | 5.920819 | UBA/TS-N domain No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)