













| General Information: |
|
| Name(s) found: |
RAD53 /
YPL153C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 821 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
DNA replication initiation
[IMP DNA repair [IMP deoxyribonucleoside triphosphate biosynthetic process [IMP nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IGI |
| Molecular Function: |
DNA replication origin binding
[IDA protein serine/threonine/tyrosine kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENITQPTQQ STQATQRFLI EKFSQEQIGE NIVCRVICTT GQIPIRDLSA DISQVLKEKR 60
61 SIKKVWTFGR NPACDYHLGN ISRLSNKHFQ ILLGEDGNLL LNDISTNGTW LNGQKVEKNS 120
121 NQLLSQGDEI TVGVGVESDI LSLVIFINDK FKQCLEQNKV DRIRSNLKNT SKIASPGLTS 180
181 STASSMVANK TGIFKDFSII DEVVGQGAFA TVKKAIERTT GKTFAVKIIS KRKVIGNMDG 240
241 VTRELEVLQK LNHPRIVRLK GFYEDTESYY MVMEFVSGGD LMDFVAAHGA VGEDAGREIS 300
301 RQILTAIKYI HSMGISHRDL KPDNILIEQD DPVLVKITDF GLAKVQGNGS FMKTFCGTLA 360
361 YVAPEVIRGK DTSVSPDEYE ERNEYSSLVD MWSMGCLVYV ILTGHLPFSG STQDQLYKQI 420
421 GRGSYHEGPL KDFRISEEAR DFIDSLLQVD PNNRSTAAKA LNHPWIKMSP LGSQSYGDFS 480
481 QISLSQSLSQ QKLLENMDDA QYEFVKAQRK LQMEQQLQEQ DQEDQDGKIQ GFKIPAHAPI 540
541 RYTQPKSIEA ETREQKLLHS NNTENVKSSK KKGNGRFLTL KPLPDSIIQE SLEIQQGVNP 600
601 FFIGRSEDCN CKIEDNRLSR VHCFIFKKRH AVGKSMYESP AQGLDDIWYC HTGTNVSYLN 660
661 NNRMIQGTKF LLQDGDEIKI IWDKNNKFVI GFKVEINDTT GLFNEGLGML QEQRVVLKQT 720
721 AEEKDLVKKL TQMMAAQRAN QPSASSSSMS AKKPPVSDTN NNGNNSVLND LVESPINANT 780
781 GNILKRIHSV SLSQSQIDPS KKVKRAKLDQ TSKGPENLQF S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
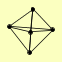
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ASF1 HIR2 HIR3 HPC2 RAD53 |
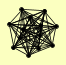
| View Details | Krogan NJ, et al. (2006) |
|
CBC2 KAP95 NUP1 NUP2 NUP60 OGG1 PCT1 RAD53 SRP1 STO1 ULP1 URB2 YRA2 |
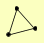
| View Details | Gavin AC, et al. (2006) |
|
HIR3 MAM33 RAD53 |
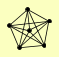
| View Details | Gavin AC, et al. (2002) |
|
ASF1 HIR3 MAM33 RAD53 RNR2 RNR4 |
| View Details | Ho Y, et al. (2002) |
|
ADH2 ASF1 CDC13 CDC16 CDC33 DUN1 EDE1 ERB1 FPR3 GID8 HHF1, HHF2 HTA2 IPP1 KAP95 KRE33 KRI1 LOC1 MDH1 MES1 MKT1 MMS4 MUS81 NHP2 NOP12 NOP2 PAA1 PTC2 PWP1 RAD53 RPC10 RPF2 SEC23 SMC3 SRP1 SWI4 TAF14 TBF1 UBI4 UTP22 YHB1 YRA1 YTA7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample 3121 run on DecaLCQ: searched for phosphorylation | Green EM, et al (2005) | |
| View Run | 3906: TAP-tagged | Green EM, et al (2005) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | sample 3829 | Green EM, et al (2005) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
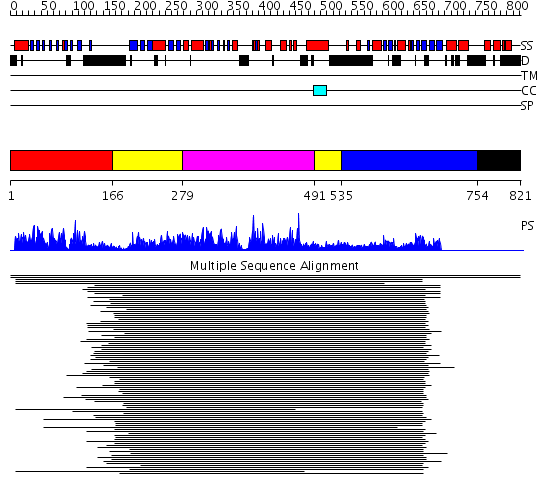
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 904.325697 | Phosphotyrosine binding domain of Rad53 | |
| 2 | View Details | [166..278] [491..534] |
622.457575 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [279..490] | 622.457575 | Calmodulin-dependent protein kinase | |
| 4 | View Details | [535..753] | 1080.0 | Phosphotyrosine binding domain of Rad53 | |
| 5 | View Details | [754..821] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)