













| General Information: |
|
| Name(s) found: |
RNR4 /
YGR180C
[SGD]
|
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 345 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA ribonucleoside-diphosphate reductase complex [IDA |
| Biological Process: |
S phase of mitotic cell cycle
[IGI DNA replication [TAS |
| Molecular Function: |
ribonucleoside-diphosphate reductase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEAHNQFLKT FQKERHDMKE AEKDEILLME NSRRFVMFPI KYHEIWAAYK KVEASFWTAE 60
61 EIELAKDTED FQKLTDDQKT YIGNLLALSI SSDNLVNKYL IENFSAQLQN PEGKSFYGFQ 120
121 IMMENIYSEV YSMMVDAFFK DPKNIPLFKE IANLPEVKHK AAFIERWISN DDSLYAERLV 180
181 AFAAKEGIFQ AGNYASMFWL TDKKIMPGLA MANRNICRDR GAYTDFSCLL FAHLRTKPNP 240
241 KIIEKIITEA VEIEKEYYSN SLPVEKFGMD LKSIHTYIEF VADGLLQGFG NEKYYNAVNP 300
301 FEFMEDVATA GKTTFFEKKV SDYQKASDMS KSATPSKEIN FDDDF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
RNR2 RNR4 |
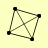
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
RNR1 RNR2 RNR3 RNR4 |
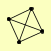
| View Details | Krogan NJ, et al. (2006) |
|
RNR1 RNR2 RNR3 RNR4 |
| View Details | Gavin AC, et al. (2006) |

|
RNR2 RNR4 |
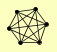
| View Details | Gavin AC, et al. (2002) |
|
ASF1 HIR3 MAM33 RAD53 RNR2 RNR4 |
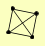
| View Details | Qiu et al. (2008) |
|
RNR1 RNR2 RNR3 RNR4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
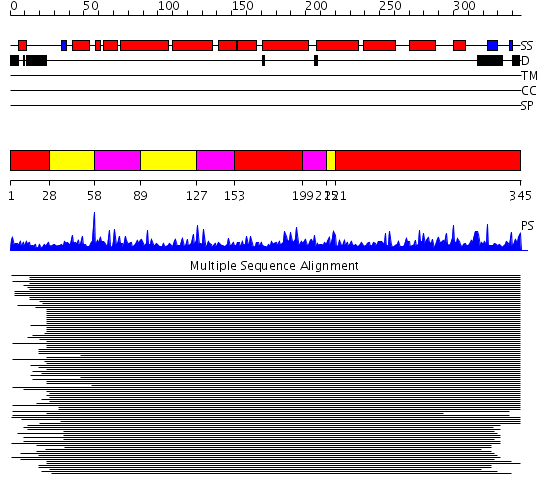
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..27] [153..198] [221..345] |
10096.522879 | Ribonucleotide reductase R2 | |
| 2 | View Details | [28..57] [89..126] [215..220] |
10096.522879 | Ribonucleotide reductase R2 | |
| 3 | View Details | [58..88] [127..152] [199..214] |
10096.522879 | Ribonucleotide reductase R2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)