













| General Information: |
|
| Name(s) found: |
MES1 /
YGR264C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 751 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA methionyl glutamyl tRNA synthetase complex [IDA |
| Biological Process: |
methionyl-tRNA aminoacylation
[IDA |
| Molecular Function: |
methionine-tRNA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFLISFDKS KKHPAHLQLA NNLKIALALE YASKNLKPEV DNDNAAMELR NTKEPFLLFD 60
61 ANAILRYVMD DFEGQTSDKY QFALASLQNL LYHKELPQQH VEVLTNKAIE NYLVELKEPL 120
121 TTTDLILFAN VYALNSSLVH SKFPELPSKV HNAVALAKKH VPRDSSSFKN IGAVKIQADL 180
181 TVKPKDSEIL PKPNERNILI TSALPYVNNV PHLGNIIGSV LSADIFARYC KGRNYNALFI 240
241 CGTDEYGTAT ETKALEEGVT PRQLCDKYHK IHSDVYKWFQ IGFDYFGRTT TDKQTEIAQH 300
301 IFTKLNSNGY LEEQSMKQLY CPVHNSYLAD RYVEGECPKC HYDDARGDQC DKCGALLDPF 360
361 ELINPRCKLD DASPEPKYSD HIFLSLDKLE SQISEWVEKA SEEGNWSKNS KTITQSWLKD 420
421 GLKPRCITRD LVWGTPVPLE KYKDKVLYVW FDATIGYVSI TSNYTKEWKQ WWNNPEHVSL 480
481 YQFMGKDNVP FHTVVFPGSQ LGTEENWTML HHLNTTEYLQ YENGKFSKSR GVGVFGNNAQ 540
541 DSGISPSVWR YYLASVRPES SDSHFSWDDF VARNNSELLA NLGNFVNRLI KFVNAKYNGV 600
601 VPKFDPKKVS NYDGLVKDIN EILSNYVKEM ELGHERRGLE IAMSLSARGN QFLQENKLDN 660
661 TLFSQSPEKS DAVVAVGLNI IYAVSSIITP YMPEIGEKIN KMLNAPALKI DDRFHLAILE 720
721 GHNINKAEYL FQRIDEKKID EWRAKYGGQQ V |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
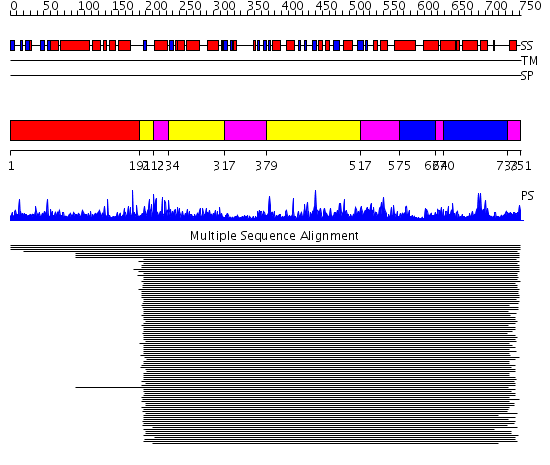
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [191..211] [234..316] [379..516] |
777.0 | Methionyl-tRNA synthetase (MetRS); Methionyl-tRNA synthetase (MetRS), Zn-domain | |
| 3 | View Details | [212..233] [317..378] [517..574] [627..639] [733..751] |
777.0 | Methionyl-tRNA synthetase (MetRS); Methionyl-tRNA synthetase (MetRS), Zn-domain | |
| 4 | View Details | [575..626] [640..732] |
777.0 | Methionyl-tRNA synthetase (MetRS); Methionyl-tRNA synthetase (MetRS), Zn-domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)