













| General Information: |
|
| Name(s) found: |
RRP6 /
YOR001W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 733 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear exosome (RNase complex)
[IDA nuclear outer membrane [TAS |
| Biological Process: |
ribosome biogenesis
[RCA mRNA catabolic process [IMP polyadenylation-dependent ncRNA catabolic process [IDA |
| Molecular Function: |
3'-5'-exoribonuclease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSENPDVLL SRVINVVRAA SSLASQDVDF YKNLDRGFSK DLKSKADKLA DMANEIILSI 60
61 DEHHESFELK EEDISDLWNN FGNIMDNLLE MSDHSLDKLN CAINSKSRGS DLQYLGEFSG 120
121 KNFSPTKRVE KPQLKFKSPI DNSESHPFIP LLKEKPNALK PLSESLRLVD DDENNPSHYP 180
181 HPYEYEIDHQ EYSPEILQIR EEIPSKSWDD SVPIWVDTST ELESMLEDLK NTKEIAVDLE 240
241 HHDYRSYYGI VCLMQISTRE RDYLVDTLKL RENLHILNEV FTNPSIVKVF HGAFMDIIWL 300
301 QRDLGLYVVG LFDTYHASKA IGLPRHSLAY LLENFANFKT SKKYQLADWR IRPLSKPMTA 360
361 YARADTHFLL NIYDQLRNKL IESNKLAGVL YESRNVAKRR FEYSKYRPLT PSSEVYSPIE 420
421 KESPWKILMY QYNIPPEREV LVRELYQWRD LIARRDDESP RFVMPNQLLA ALVAYTPTDV 480
481 IGVVSLTNGV TEHVRQNAKL LANLIRDALR NIKNTNEEAT PIPSSETKAD GILLETISVP 540
541 QIRDVMERFS VLCNSNISKS RAKPVTNSSI LLGKILPREE HDIAYSKDGL PNKVKTEDIR 600
601 IRAQNFKSAL ANLEDIIFEI EKPLVVPVKL EEIKTVDPAS APNHSPEIDN LDDLVVLKKK 660
661 NIQKKQPAKE KGVTEKDAVD YSKIPNILSN KPGQNNRQQK KRRFDPSSSD SNGPRAAKKR 720
721 RPAAKGKNLS FKR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
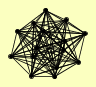
|
CSL4 DIS3 LRP1 MPP6 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 SRP1 |
| View Details | Krogan NJ, et al. (2006) |
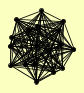
|
CSL4 DDI1 DIS3 ENT3 LRP1 MPP6 MTR3 OSM1 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 YBL055C |
| View Details | Gavin AC, et al. (2006) |
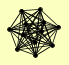
|
CSL4 DIS3 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 |
| View Details | Gavin AC, et al. (2002) |

|
CMD1 CSL4 CTR9 DIS3 ECM16 GCN1 LRP1 MAM33 MLC1 MTR3 MYO2 NMD5 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SAM1 SHE4 SKI2 SKI3 SKI6 SKI7 SRP1 UTP10 UTP20 UTP22 YIR035C |
| View Details | Ho Y, et al. (2002) |

|
DIS3 EAF3 FIP1 HAS1 HPR1 KAP95 MES1 MFT1 NAM8 NOP6 NUP1 NUP2 NUP60 PAP1 PCT1 REB1 RNT1 RRP4 RRP43 RRP6 RTT103 SIF2 SIN3 SNU56 SRP1 STO1 THO2 TRA1 UME1 YPR090W |
| View Details | Qiu et al. (2008) |
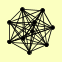
|
CSL4 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
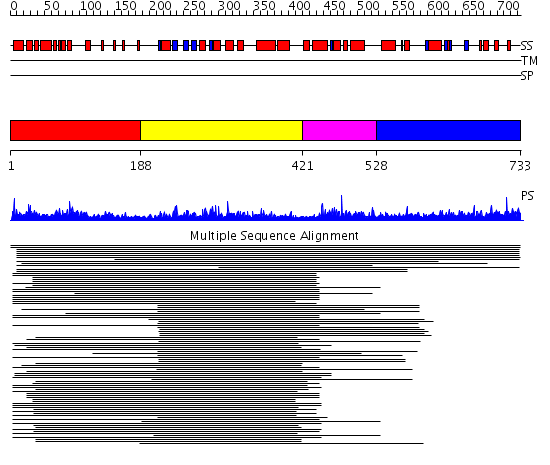
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 8.69897 | No description for 1j5fA_ was found. | |
| 2 | View Details | [188..420] | 1049.09691 | No description for 1m0yA_ was found. | |
| 3 | View Details | [421..527] | 11.125993 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [528..733] | 1.030973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)