













| General Information: |
|
| Name(s) found: |
RRP42 /
YDL111C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 265 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasmic exosome (RNase complex)
[IDA nuclear exosome (RNase complex) [IDA nuclear outer membrane [TAS |
| Biological Process: |
mRNA catabolic process
[IPI |
| Molecular Function: |
3'-5'-exoribonuclease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSVAEKSY LYDSLASTPS IRPDGRLPHQ FRPIEIFTDF LPSSNGSSRI IASDGSECIV 60
61 SIKSKVVDHH VENELLQVDV DIAGQRDDAL VVETITSLLN KVLKSGSGVD SSKLQLTKKY 120
121 SFKIFVDVLV ISSHSHPVSL ISFAIYSALN STYLPKLISA FDDLEVEELP TFHDYDMVKL 180
181 DINPPLVFIL AVVGNNMLLD PAANESEVAN NGLIISWSNG KITSPIRSVA LNDSNVKSFK 240
241 PHLLKQGLAM VEKYAPDVVR SLENL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
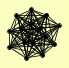
|
CSL4 DIS3 LRP1 MPP6 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 SRP1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
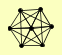
|
DIS3 RRP4 RRP42 RRP43 RRP45 RRP46 SKI6 |
| View Details | Krogan NJ, et al. (2006) |
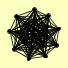
|
CSL4 DDI1 DIS3 ENT3 LRP1 MPP6 MTR3 OSM1 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 YBL055C |
| View Details | Gavin AC, et al. (2006) |
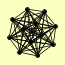
|
CSL4 DIS3 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 |
| View Details | Gavin AC, et al. (2002) |

|
CMD1 CSL4 CTR9 DIS3 ECM16 GCN1 LRP1 MAM33 MLC1 MTR3 MYO2 NMD5 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SAM1 SHE4 SKI2 SKI3 SKI6 SKI7 SRP1 UTP10 UTP20 UTP22 YIR035C |
| View Details | Ho Y, et al. (2002) |
|
APA1 GAR1 LSM2 LSM8 QCR2 RPN12 RPN8 RRP42 SMB1 TIF6 YGL117W |
| View Details | Qiu et al. (2008) |
|
CSL4 DIS3 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
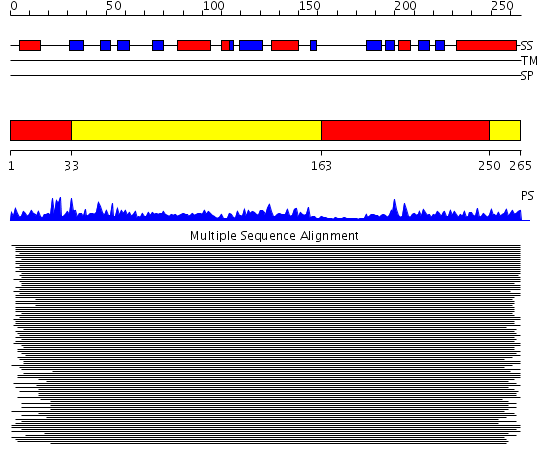
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..32] [163..249] |
47.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [33..162] [250..265] |
47.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)