













| General Information: |
|
| Name(s) found: |
NMD5 /
YJR132W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1048 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
protein import into nucleus
[IMP |
| Molecular Function: |
protein transmembrane transporter activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDITELLQCF ACTLDHNAAV RTNAETHLKN ASKVPGFLGA CLDIIAADEV PENIKLSASL 60
61 YFKNKITYGW SAGARQGSNE LLDSHVDPDE KPVVKDMLIK TMVSVSKTSP RCIRVLKSAL 120
121 TVIISEDYPS KKWGNLLPNS LELLANEDIT VTYVGLLCLA EIFRTYRWKN NDERQDLEEL 180
181 ILNYFPALLN YGANVLFQDG KYMNNEQIGE LVKLIIKIYK FVSYHDLPFT LQRSESFTPW 240
241 ACFFVSIIQQ PLPQEVLSIS DIEVRSKNPW VKCKKWALAN LYRLFQRYAS TSLTRKFQYD 300
301 EFKQMYCEEF LTQFLQVVFD QIEKWGTGQL WLSDECLYYI LNFVEQCVVQ KTTWKLVGPH 360
361 YNVILQHVIF PLLKPTAETL EAFDNDPQEY INRNMDFWDV GYSPDLAALA LLTTCVTKRG 420
421 KTTLQPTLEF MVSTLQSAVG DYNNIMLDNA LQIESCLRIF SSIIDRLITK DSPFASEMEK 480
481 FILTYVLPFF KSQYGFLQSR VCDICSKLGS MDFKDPVITS TIYEGVMNCL NNSSNSLPVE 540
541 LTAALALQTF ISDDQFNMKL SEHVVPTMQK LLSLSNDFES DVISGVMQDF VEQFAEQLQP 600
601 FGVELMNTLV QQFLKLAIDL HETSNLDPDS FTNVDSIPDE SDKQMAALGI LSTTISILLS 660
661 FENSPEILKN LEQSFYPAAE FILKNDIEDF YRECCEFVEN STFLLRDITP ISWKILELIG 720
721 ECNRKPDSMV SYYLSDFMLA LNNILIYGRN ELKKNEFYTK IIFEIYQKAV TAEDNSLDDL 780
781 RVVFDLSQEL VLALDDSLPQ QYRERLLADV VGSILTQKNE LKTNVVFSVT AFNVVISNMI 840
841 TEPLITLQYL KQQGCLEIFF QTWITDYIPN YKRCYDIKLS VLALLKIILK LESNDYSVLN 900
901 LENLVPQLGS IVTQLASRLP TALRQLANQR KEFSSSGFEE DTKWDENFLD VGDDDENDDE 960
961 GDLTEKYLEL IKNRADSLDF VDGYDAKETF DDLEEDPLTG SILDTVDVYK VFKESIANLQ1020
1021 HVDSNRYQGI LRHLTPADQE LFMGIMNA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
ECM5 MDN1 MTR10 NMD5 TUB2 USA1 |
| View Details | Gavin AC, et al. (2002) |
|
BDF2 CDC55 CKB1 CMD1 CSL4 CTR9 DIS3 DYN1 ECM16 ECM5 GCN1 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MAM33 MDN1 MIS1 MLC1 MSB1 MTR3 MYO2 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 RSC3 SAM1 SHE4 SKI6 SKI7 SRP1 TEL1 TSC11 USO1 UTP10 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |

|
AAC3 AHA1 AIM46 ARP2 ARP4 ASA1 ATG17 ATG29 ATP3 CDC33 CIC1 CIS1 CPR6 CYS4 FAA4 FET3 FOL2 GCD1 GCD11 GCD2 GCD6 GCD7 GCN1 GCN3 GRH1 GSP1 GSP2 HAP2 HAP5 HXT7 IDH1 IPP1 KAP95 LEM3 LOC1 LOS1 MAM33 MET18 MTQ1 NAP1 NMD5 NOP16 NSA2 NUS1 PET10 PHO84 PMA1 POL5 PPH21 PPH22 PRB1 PRO3 PSE1 REP1 RHR2 RPN8 SAH1 SAP190 SCW4 SDH2 SPE3 SPS1 SSK2 SUI3 TCP1 TIF1, TIF2 TIF6 VAC8 VMA8 VPS74 YHR033W YLR326W YRA1 YRA2 YTM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
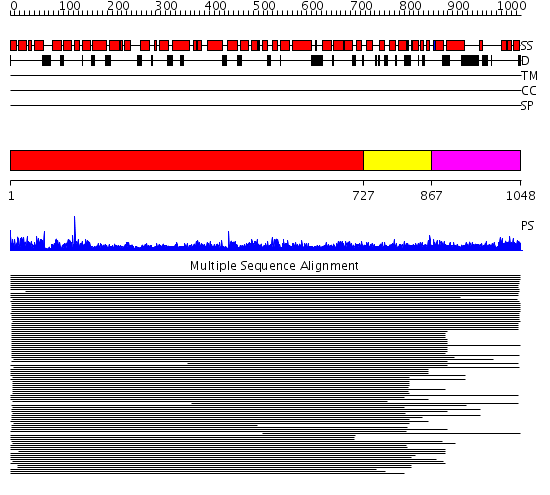
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..726] | 11.125995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [727..866] | 2.097994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [867..1048] | 1.055982 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)