













| General Information: |
|
| Name(s) found: |
VTH2 /
YJL222W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1549 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
Golgi to vacuole transport
[ISS |
| Molecular Function: |
signal sequence binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALFRALYII WVFLLIPLSN AEEFTPKVTR TLSRYVFDIV NFDDSNTLIR AEEDSVEISF 60
61 DAGENWKTID EIEEPIESFV VDPFRGHDRA FAFVKTAPKF YVTDDQGKSW RPLTIPISEK 120
121 ASNYFCDVTT HPIKKKHLII RCDLLTIKNS GLMYVGREIY TTNDGVSFSQ VKPSFGKIDG 180
181 HISTARCDFI KSSEDSDLGG NDASILCLFR NTEYIESTGS TIDKSELILS ADGGETFKEL 240
241 VQFKDKVVSR YEILKHHVIV LTQDDMYNEM SSTNIWISND VSTFQVARTP TKIRHVNMGQ 300
301 IHEDSIGRIV LPVSRERDDE DSNQPGAAEV LISDSEGLKF LPINWIPNNQ FGYINVAYPG 360
361 FLKGTFFGSF HPFIEYSDRK RKYSRQKVRE ETKVSVDNGL TWTNLKVVDR ENVDLFGCDV 420
421 TKPERCSLQT HFYDLRNLNP SAGIMMISGI VGDGSAYNWK EEKTFISRDS GLTWRLVHNS 480
481 TGLYTTGDLG NIIMYIPYRS NENGDVPSKF YYSLDQGKTW GEYDLIMPIY PYRLVSTISD 540
541 GSGSKFILTG TSITEDPIFI TYSIDFSAVF DYKSCEEGDF EDWNLADGKC VNGAKYKYRR 600
601 RKQDAQCLVK KAFKDLSLDE TPCNSCTGSD YECSFEFVRD AKGDCIPDYN LIALSDICDK 660
661 SKGKSVLVKP LQLIKGDKCK TPMKIESVDI PCDEIPKEGS SDKEIVTTEN KFDFEIKFYQ 720
721 YFDTVADESL VMLNSIGDAY ISHDGGQTIK RFDTDGEKIV EIVFNPYFNS SAYLFGSKGN 780
781 IFLTHDRGYS FMIAKLPEAR QLGMPLDFSA KAQDTFIYYG GKNCESILSP ECHAVAYLTK 840
841 DGGETFTEML DNAIHCEFAG TLFKYPSNDD MVMCQVKEKF SQTRSLVSST DFFQDDRKTV 900
901 FENIIGYLST GGYIIVAVPH EDNELRAYVT NDGAEFTEAK FPYDEDIGKQ DAFTILGSEE 960
961 GSIFLHLATN LESGHDFGNL LKSNSNGTSF VTLEHAVNRN TFGYVDFEKV QGLEGIIITN1020
1021 IVSNSEKVGE NKEDEQLKTK ITFNDGSDWN FLKPPKKDSE GKKFPCDSVS LDKCSLHLHG1080
1081 YTERKDIRDT YSSGSALGMM FGVGNVGDRL LPYEECSTFL TTDGGETWTE VKKGPHQWEY1140
1141 GDHGGVLVLV PENAETDSIS YSTDFGKTWK DYKFCGDKVL VKDIITVPRD SALRFLLFGE1200
1201 AKNMGSGSFR TYTIDFRNIF ERQCEFDITG RKRADFKYSP LGSRTGCLFG HKTEFLRKTD1260
1261 EKCFIGNIPL SEFSRNVKNC PCTRQDFECD YNFYKASDGT CKLVKGLSSA NGADICKKEP1320
1321 DLIEYYDSSG YRKIPLSTCK GGLKLDAHLA PHPCPGKEKA FREKYSINTG AYALVFVTIL1380
1381 LVIFFVAWFV YDRGIRRNGG FSRFEEIRLG DDGLIENNRT DRVVNIIVRL GLCISLITKS1440
1441 AFQRAKAGTA QLSSKFRARF GNKKGATYSS LLHDQLSDEP DGFHEDSNDL SSFRGQGSNS1500
1501 EIEQEDVDTS QQEHTSRTDL LGASNIPDAL PARSASHESD LAAARSEDK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | 3906: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
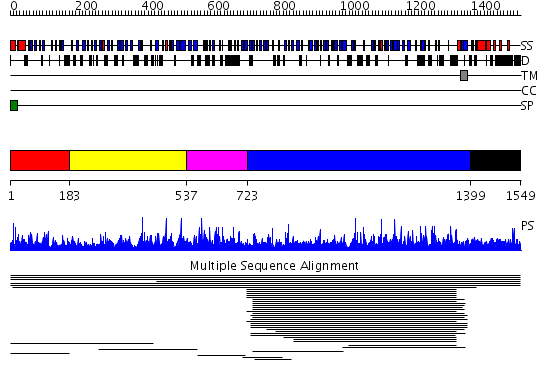
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 1.005938 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [183..536] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [537..722] | 1.007947 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [723..1398] | 7.017001 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1399..1549] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|