













| General Information: |
|
| Name(s) found: |
ECM5 /
YMR176W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1411 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
cellular cell wall organization
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGHDSVTKI SHILNEPVNE KVMVQNGFHE SSKIADIELE IQERPSIKQW ESPRSAVIPT 60
61 SNHNFSPFLY TQFKSRGAAP FAPETIKSVD LVELPEGVPA RVFHEKTGLF YQISPHSIPT 120
121 FILAKKELPD PIKFYELVED LGSVYGCVKL KIIPDADKFT QLNVDVDRLW FKARKQFFNS 180
181 NEFQRTKIVD FYAKLYNFHN KIKKSTLTRI PSIDKRTLDL YRLRSCVKLR GGFNAVCEKK 240
241 LWAQIGRELG YSGRIMSSLS TSLRSAYAKI LLDFDIYEEE EQAARNNEKN EDMVESEIFR 300
301 HSNSRSRDEE EPLHKKAKIH RDVFRAGSIN HEFKRMRDIK HIKGFPTYFN SLTEFKLGYT 360
361 QSTETTLPGY DFTFWENGME IYDKSKYETK TSPVYNLRQY YEKSLAVFTA IVAKFGSSYP 420
421 DLFAKHTTLP QKEFERLYFH LLSEHFIDFE IDTGLGLPCS MRSPGNNSSN EKFAIKNILD 480
481 QWNLDNIPLN ELSLLQHLDL DMANFTRTTY DIGMLFSCQG WSVSDHFLPS IDFNHLGSTK 540
541 LVYSIAPKDM EKFEALIARG KSEWDTIQSR PRYSTSDDEL KSFIETDFYK SFLDAEQSAD 600
601 YSNTGDNSKN SFPEDKIAGN TLHDGSQSDF IFEPNFILAN GIKLYKTTQE QGSYIFKFPK 660
661 AFTCSIGSGF YLSQNAKFAP SSWLRFSSEA AKWTSKMGFL PGLDVNQLLI NALLNSNNPV 720
721 LRKKCRDLIS NYVVEEAENS KKLGELIGTV DVVYNKLNYI SDISLESTGL SKIVVTHGAL 780
781 QRNLSLKEFV VLLEKPENGA HSICGIPIRD QSGNLNVCLH SYFDSASLGI ALDGLDKPPT 840
841 SYLLVHNEDF EKKWDVLMTS TFRNRTVPLN IIQYLISHTD SNTEFNRMLR SNFDDSLLLI 900
901 EKCKKFIKTF VDVSCSVKDV DFGNGFNLRH LPLKFSDNMA DNLESLYESV RKCSIEFSEK 960
961 PTIIRLYHVS RQFPIDNRDI IDGNNLDLLK ELYQKSLTIP LKVSYWTKLT RKICRLEWLS1020
1021 VYEHIFIERC DIKNEDPAKY SLPLLYSYFE FGLKYCDSED IDKLGEVRKL ILKYQDMMQK1080
1081 VRVFLKKDPP SKISLSDLED VLLDIEEYRL PIQSSFFSEL DYVIREIENA KKMNDVNILY1140
1141 NTDNIDKIDE LIRKNDPKFV KFANQFNGSR LDKRPLASDN SGSVKAKQEL KVFKLWNQHL1200
1201 DQIMQKNKFI EILPSIFRCL DLKSDKYIPL ESCSKRQTKY CFCRRVEEGT AMVECEICKE1260
1261 WYHVDCISNG ELVPPDDPNV LFVCSICTPP CMAVDNIEGV TFELDDLKRI LVESLKLSLI1320
1321 PDPPILKNLF DVFAFALNFK NEMEKELFTN GYVNQLSSTH KIKYYLRKLK GSQCGFTNLT1380
1381 DPLRKHCQVK DAEAIKWLTD NGRIIITGIP N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
ECM5 IML1 NMD5 PSD2 PSH1 YFR006W |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
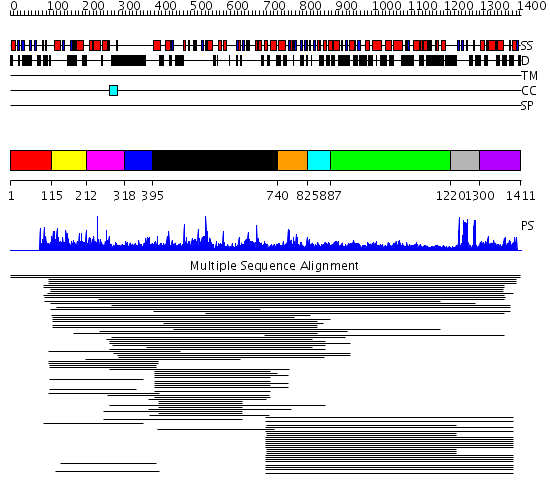
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [115..211] | 91.0103 | DNA-binding domain from the dead ringer protein | |
| 3 | View Details | [212..317] | 91.0103 | DNA-binding domain from the dead ringer protein | |
| 4 | View Details | [318..394] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [395..739] | 9.11995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [740..824] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [825..886] | 4.09691 | Interferon-induced guanylate-binding protein 1 (GBP1), C-terminal domain; Interferon-induced guanylate-binding protein 1 (GBP1), N-terminal domain | |
| 8 | View Details | [887..1219] | 23.30103 | Heavy meromyosin subfragment | |
| 9 | View Details | [1220..1299] | 2.30103 | Nuclear corepressor KAP-1 (TIF-1beta) | |
| 10 | View Details | [1300..1411] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)