













| General Information: |
|
| Name(s) found: |
PSH1 /
YOL054W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 406 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
RNA elongation from RNA polymerase II promoter
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDELHNRLL HQNDGTKDAI LYKIIESLVC SICHDYMFVP MMTPCGHNYC YGCLNTWFAS 60
61 NTQKELACPQ CRSDITTIPA LNTTLQQYLS FILEKLRDQN DESFKKLLTT KTKEENDYKN 120
121 DKEKDTLFDK VFKNSALAVA DDSDDGITRC SNCHWELDPD EVEDGNVCPH CNARIRNYAG 180
181 GRDEFDEEEY SEGELDEIRE SMRRRRENRF ASTNPFANRD DVSSEDDDSS EEEPMREHIP 240
241 LGRWARSHNR SIAVDAVDDE DDEEEDEEEE EEMDSDLKDF IEDDEDDEDE DGSRRNLVLS 300
301 ALKNRHVIIT DDEEEEQRRH ATEEEDRDSD FYEHNDDGFV SGDSLDEDQK EVTRIQSSSD 360
361 SEDRSLSYSG SSDVKDNNDD NTEELDDPQP KRQKRFRVVL GDSDDE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
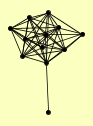
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ASF1 CKA1 HAT1 HAT2 HHF1, HHF2 HHT1, HHT2 HIF1 MAM33 MCM22 POB3 PSH1 SPT16 |
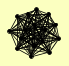
| View Details | Krogan NJ, et al. (2006) |
|
ARA2 AYT1 BRO1 CDC73 CTR9 GLO4 HHT1, HHT2 LEO1 PAF1 PML39 POB3 PSH1 RTF1 SEN1 SPT16 YHR035W |
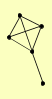
| View Details | Gavin AC, et al. (2006) |
|
ECM5 PSD2 PSH1 RPL38 YFR006W |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |

|
EDE1 GAC1 HHF1, HHF2 HTA1 HTA2 HTB1 HTB2 KNS1 MAM33 NOB1 POB3 PSH1 SPT16 SYP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
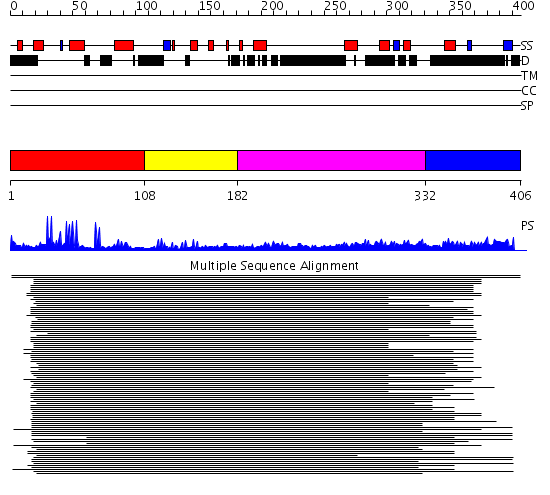
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 35.522879 | Cbl; N-terminal domain of cbl (N-cbl); CBL | |
| 2 | View Details | [108..181] | 2.30103 | Nuclear factor XNF7 | |
| 3 | View Details | [182..331] | 3.250986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [332..406] | 16.522879 | TNF receptor associated factor 3 (TRAF3); TRAF3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)