













| General Information: |
|
| Name(s) found: |
USO1 /
YDL058W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[IMP protein complex assembly [IDA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDIIQGLIQQ PKIQSVDETI PTLCDRVENS TLISDRRSAV LGLKAFSRQY RESVIASGLK 60
61 PLLNTLKRDY MDEDSVKAIL ETILILFIRG DGHDDLTRGW ISQQSRLQNG KYPSPLVMKQ 120
121 EKEQVDQFSL WIADALTQSE DLIHLLVEFW EIDNFHIRLY TIQLLEAVMA TRPLKARSAL 180
181 ISLPTSISTM VSLLDDMHEP IRDEAILLLM AVVNDSPHVQ KLVAFENIFE RLFSIIEEEG 240
241 GLRGSLVVND CLSLINNILK YNTSNQTLFL ETGNLPKLAH LLSEPISQDE VFFWNDQRIV 300
301 NINTALDIVS LTVEPGNTVT TKHQNALLDS SVLMVVLRLA FFHNIPKKVR PVALLTAANM 360
361 VRSNEHAQLE FSKIDVPYFD PSLPVNSTAN GGPIKLIPVV SILINWMLYA NSVHTFDTRV 420
421 ACSRLLKAYF MDNFDLQRDF LLKQVQLCNN STNNVGDNAK ENGGSNKSDK ESDSDKDTDG 480
481 KDGTEYEGSF KANLFEVLLN YDAELNLNPF KLFFTTDIFM FFFQQDHKYS EELREITRNV 540
541 TTGNDLEDEE PLKAIQTISE LLTTSLTAAD IRIPISYLTF LIYWLFGDFK ATNDFLSDKS 600
601 VIKSLLSFSY QIQDEDVTIK CLVTMLLGVA YEFSSKESPF PRKEYFEFIT KTLGKDNYAS 660
661 RIKQFKKDSY FSKVDMNEDS ILTPELDETG LPKVYFSTYF IQLFNENIYR IRTALSHDPD 720
721 EEPINKISFE EVEKLQRQCT KLKGEITSLQ TETESTHENL TEKLIALTNE HKELDEKYQI 780
781 LNSSHSSLKE NFSILETELK NVRDSLDEMT QLRDVLETKD KENQTALLEY KSTIHKQEDS 840
841 IKTLEKGLET ILSQKKKAED GINKMGKDLF ALSREMQAVE ENCKNLQKEK DKSNVNHQKE 900
901 TKSLKEDIAA KITEIKAINE NLEEMKIQCN NLSKEKEHIS KELVEYKSRF QSHDNLVAKL 960
961 TEKLKSLANN YKDMQAENES LIKAVEESKN ESSIQLSNLQ NKIDSMSQEK ENFQIERGSI1020
1021 EKNIEQLKKT ISDLEQTKEE IISKSDSSKD EYESQISLLK EKLETATTAN DENVNKISEL1080
1081 TKTREELEAE LAAYKNLKNE LETKLETSEK ALKEVKENEE HLKEEKIQLE KEATETKQQL1140
1141 NSLRANLESL EKEHEDLAAQ LKKYEEQIAN KERQYNEEIS QLNDEITSTQ QENESIKKKN1200
1201 DELEGEVKAM KSTSEEQSNL KKSEIDALNL QIKELKKKNE TNEASLLESI KSVESETVKI1260
1261 KELQDECNFK EKEVSELEDK LKASEDKNSK YLELQKESEK IKEELDAKTT ELKIQLEKIT1320
1321 NLSKAKEKSE SELSRLKKTS SEERKNAEEQ LEKLKNEIQI KNQAFEKERK LLNEGSSTIT1380
1381 QEYSEKINTL EDELIRLQNE NELKAKEIDN TRSELEKVSL SNDELLEEKQ NTIKSLQDEI1440
1441 LSYKDKITRN DEKLLSIERD NKRDLESLKE QLRAAQESKA KVEEGLKKLE EESSKEKAEL1500
1501 EKSKEMMKKL ESTIESNETE LKSSMETIRK SDEKLEQSKK SAEEDIKNLQ HEKSDLISRI1560
1561 NESEKDIEEL KSKLRIEAKS GSELETVKQE LNNAQEKIRI NAEENTVLKS KLEDIERELK1620
1621 DKQAEIKSNQ EEKELLTSRL KELEQELDST QQKAQKSEEE RRAEVRKFQV EKSQLDEKAM1680
1681 LLETKYNDLV NKEQAWKRDE DTVKKTTDSQ RQEIEKLAKE LDNLKAENSK LKEANEDRSE1740
1741 IDDLMLLVTD LDEKNAKYRS KLKDLGVEIS SDEEDDEEDD EEDEEEGQVA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
GLN4 USO1 |
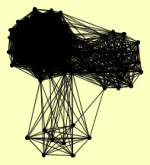
| View Details | Gavin AC, et al. (2002) |
|
BDF2 CDC55 CKB1 CLU1 CRN1 DYN1 ECM29 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MEC1 MIP6 MIS1 MRPL23 MSB1 NAB3 NAB6 NIP1 NMD5 NRD1 PFK1 POL1 POL12 PRI1 PRI2 PRO1 PRT1 PSD2 PSE1 PSH1 RFC1 RFC3 RFC4 RFC5 ROM2 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 SEN54 SUI3 SUP35 SUP45 TAO3 TEL1 TSC11 USO1 UTP20 VAC14 VPS5 VTH2 YFR006W YJL017W YPR077C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
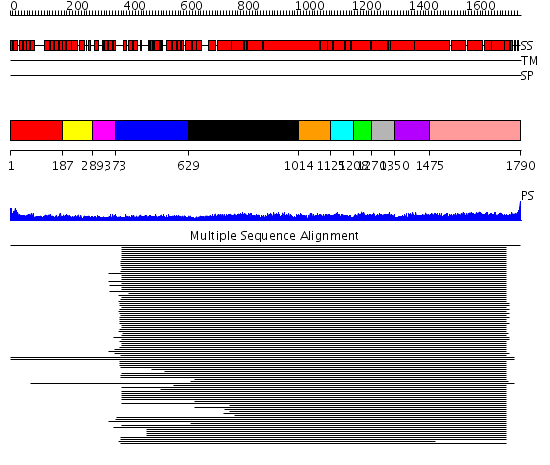
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | 14.86 | beta-Catenin | |
| 2 | View Details | [187..288] | 14.86 | beta-Catenin | |
| 3 | View Details | [289..372] | 14.86 | beta-Catenin | |
| 4 | View Details | [373..628] | 137.201249 | Heavy meromyosin subfragment | |
| 5 | View Details | [629..1013] | 137.201249 | Heavy meromyosin subfragment | |
| 6 | View Details | [1014..1124] | 9.09691 | Tropomyosin | |
| 7 | View Details | [1125..1207] | 9.09691 | Tropomyosin | |
| 8 | View Details | [1208..1269] | 9.09691 | Tropomyosin | |
| 9 | View Details | [1270..1349] | 9.09691 | Tropomyosin | |
| 10 | View Details | [1350..1474] | 11.85 | Heavy meromyosin subfragment | |
| 11 | View Details | [1475..1790] | 11.85 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)