













| General Information: |
|
| Name(s) found: |
GLN4 /
YOR168W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC |
| Biological Process: |
glutaminyl-tRNA aminoacylation
[IMP |
| Molecular Function: |
glutamine-tRNA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVEELTQL FSQVGFEDKK VKEIVKNKKV SDSLYKLIKE TPSDYQWNKS TRALVHNLAS 60
61 FVKGTDLPKS ELIVNGIING DLKTSLQVDA AFKYVKANGE ASTKMGMNEN SGVGIEITED 120
121 QVRNYVMQYI QENKERILTE RYKLVPGIFA DVKNLKELKW ADPRSFKPII DQEVLKLLGP 180
181 KDERDLIKKK TKNNEKKKTN SAKKSSDNSA SSGPKRTMFN EGFLGDLHKV GENPQAYPEL 240
241 MKEHLEVTGG KVRTRFPPEP NGYLHIGHSK AIMVNFGYAK YHNGTCYLRF DDTNPEKEAP 300
301 EYFESIKRMV SWLGFKPWKI TYSSDYFDEL YRLAEVLIKN GKAYVCHCTA EEIKRGRGIK 360
361 EDGTPGGERY ACKHRDQSIE QNLQEFRDMR DGKYKPGEAI LRMKQDLNSP SPQMWDLIAY 420
421 RVLNAPHPRT GTKWRIYPTY DFTHCLVDSM ENITHSLCTT EFYLSRESYE WLCDQVHVFR 480
481 PAQREYGRLN ITGTVLSKRK IAQLVDEKFV RGWDDPRLFT LEAIRRRGVP PGAILSFINT 540
541 LGVTTSTTNI QVVRFESAVR KYLEDTTPRL MFVLDPVEVV VDNLSDDYEE LATIPYRPGT 600
601 PEFGERTVPF TNKFYIERSD FSENVDDKEF FRLTPNQPVG LIKVSHTVSF KSLEKDEAGK 660
661 IIRIHVNYDN KVEEGSKPKK PKTYIQWVPI SSKYNSPLRV TETRVYNQLF KSENPSSHPE 720
721 GFLKDINPES EVVYKESVME HNFGDVVKNS PWVVDSVKNS EFYVEEDKDS KEVCRFQAMR 780
781 VGYFTLDKES TTSKVILNRI VSLKDATSK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
GLN4 USO1 |
| View Details | Krogan NJ, et al. (2006) |
|
GLN4 PBI2 PHO4 PRB1 SOD2 YNL019C |
| View Details | Gavin AC, et al. (2006) |
|
GLN4 SSL2 YOR352W |
| View Details | Qiu et al. (2008) |
|
GLN4 RKI1 RPL24A RPL24B |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
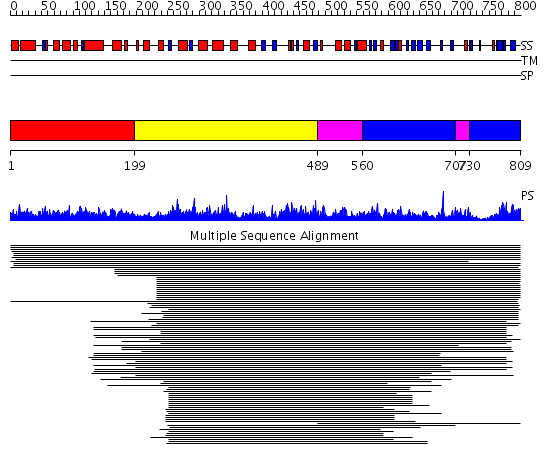
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [199..488] | 1306.0 | Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain; Glutaminyl-tRNA synthetase (GlnRS) | |
| 3 | View Details | [489..559] [707..729] |
1306.0 | Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain; Glutaminyl-tRNA synthetase (GlnRS) | |
| 4 | View Details | [560..706] [730..809] |
1306.0 | Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain; Glutaminyl-tRNA synthetase (GlnRS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)