













| General Information: |
|
| Name(s) found: |
VAC14 /
YLR386W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 880 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA vacuole [IDA |
| Biological Process: |
vacuole inheritance
[IMP phospholipid metabolic process [IGI |
| Molecular Function: |
enzyme activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKSIAKGLS DKLYEKRKAA ALELEKLVKQ CVLEGDYDRI DKIIDELCRD YAYALHQPMA 60
61 RNAGLMGLAA TAIALGINDV GRYLRNILPP VLACFGDQND QVRFYACESL YNIAKIAKGE 120
121 ILVYFNEIFD VLCKISADTE NSVRGAAELL DRLIKDIVAE RASNYISIVN NGSHGLLPAI 180
181 KTDPISGDVY QEEYEQDNQL AFSLPKFIPL LTERIYAINP DTRVFLVDWL KVLLNTPGLE 240
241 LISYLPSFLG GLFTFLGDSH KDVRTVTHTL MDSLLHEVDR ISKLQTEIKM KRLERLKMLE 300
301 DKYNNSSTPT KKADGALIAE KKKTLMTALG GLSKPLSMET DDTKLSNTNE TDDERHLTSQ 360
361 EQLLDSEATS QEPLRDGEEY IPGQDINLNF PEVITVLVNN LASSEAEIQL IALHWIQVIL 420
421 SISPNVFIPF LSKILSVLLK LLSDSDPHIT EIAQLVNGQL LSLCSSYVGK ETDGKIAYGP 480
481 IVNSLTLQFF DSRIDAKIAC LDWLILIYHK APNQILKHND SMFLTLLKSL SNRDSVLIEK 540
541 ALSLLQSLCS DSNDNYLRQF LQDLLTLFKR DTKLVKTRAN FIMRQISSRL SPERVYKVIS 600
601 SILDNYNDTT FVKMMIQILS TNLITSPEMS SLRNKLRTCE DGMFFNSLFK SWCPNPVSVI 660
661 SLCFVAENYE LAYTVLQTYA NYELKLNDLV QLDILIQLFE SPVFTRMRLQ LLEQQKHPFL 720
721 HKCLFGILMI IPQSKAFETL NRRLNSLNIW TSQSYVMNNY IRQRENSNFC DSNSDISQRS 780
781 VSQSKLHFQE LINHFKAVSE EDEYSSDMIR LDHGANNKSL LLGSFLDGID EDKQEIVTPI 840
841 SPMNEAINEE MESPNDNSSV ILKDSGSLPF NRNVSDKLKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
FIG4 VAC14 |
| View Details | Krogan NJ, et al. (2006) |
|
FIG4 KRE5 OAF1 VAC14 |
| View Details | Gavin AC, et al. (2006) |
|
MEC1 SEN54 VAC14 |
| View Details | Gavin AC, et al. (2002) |
|
ACC1 CCT5 CLU1 CRN1 ECM29 KAP123 MEC1 MIP6 MIS1 MRPL23 NAB3 NAB6 NRD1 ROM2 RPA135 SEN54 SIT4 SUI3 SUP35 SUP45 TAO3 USO1 UTP20 VAC14 VPS5 YJL017W |
| View Details | Ho Y, et al. (2002) |

|
BFR2 BIR1 BNA5 CDC7 COR1 DOG1 ECM10 ENP1 FET4 FPR3 HAS1 JIP3 MAM33 NUT1 PDC5 PDC6 PIM1 PRP8 PST2 QCR2 RPC19 SAP185 SAP190 SAR1 SEC27 SIT4 SNP1 SRP1 STI1 THI3 TPS1 UBI4 VAC14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
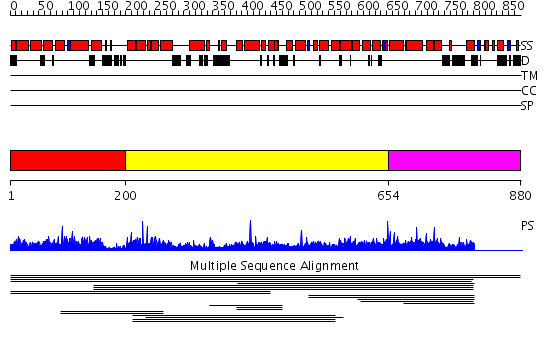
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [200..653] | 11.67 | Clathrin heavy chain proximal leg segment | |
| 3 | View Details | [654..880] | 1.009959 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [787..880] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)