













| General Information: |
|
| Name(s) found: |
PIM1 /
YBL022C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1133 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
response to heat
[IEP proteolysis [IGI |
| Molecular Function: |
ATP-dependent peptidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRTRTTKTL STVARTTRAI QYYRSIAKTA AVSQRRFAST LTVRDVENIK PSHIIKSPTW 60
61 QEFQHQLKDP RYMEHFAQLD AQFARHFMAT NSGKSILAKD DSTSQKKDED VKIVPDEKDT 120
121 DNDVEPTRDD EIVNKDQEGE ASKNSRSSAS GGGQSSSSRS DSGDGSSKQK PPKDVPEVYP 180
181 QMLALPIARR PLFPGFYKAV VISDERVMKA IKEMLDRQQP YIGAFMLKNS EEDTDVITDK 240
241 NDVYDVGVLA QITSAFPSKD EKTGTETMTA LLYPHRRIKI DELFPPNEEK EKSKEQAKDT 300
301 DTETTVVEDA NNPEDQESTS PATPKLEDIV VERIPDSELQ HHKRVEATEE ESEELDDIQE 360
361 GEDINPTEFL KNYNVSLVNV LNLEDEPFDR KSPVINALTS EILKVFKEIS QLNTMFREQI 420
421 ATFSASIQSA TTNIFEEPAR LADFAAAVSA GEEDELQDIL SSLNIEHRLE KSLLVLKKEL 480
481 MNAELQNKIS KDVETKIQKR QREYYLMEQL KGIKRELGID DGRDKLIDTY KERIKSLKLP 540
541 DSVQKIFDDE ITKLSTLETS MSEFGVIRNY LDWLTSIPWG KHSKEQYSIP RAKKILDEDH 600
601 YGMVDVKDRI LEFIAVGKLL GKVDGKIICF VGPPGVGKTS IGKSIARALN RKFFRFSVGG 660
661 MTDVAEIKGH RRTYIGALPG RVVQALKKCQ TQNPLILIDE IDKIGHGGIH GDPSAALLEV 720
721 LDPEQNNSFL DNYLDIPIDL SKVLFVCTAN SLETIPRPLL DRMEVIELTG YVAEDKVKIA 780
781 EQYLVPSAKK SAGLENSHVD MTEDAITALM KYYCRESGVR NLKKHIEKIY RKAALQVVKK 840
841 LSIEDSPTSS ADSKPKESVS SEEKAENNAK SSSEKTKDNN SEKTSDDIEA LKTSEKINVS 900
901 ISQKNLKDYV GPPVYTTDRL YETTPPGVVM GLAWTNMGGC SLYVESVLEQ PLHNCKHPTF 960
961 ERTGQLGDVM KESSRLAYSF AKMYLAQKFP ENRFFEKASI HLHCPEGATP KDGPSAGVTM1020
1021 ATSFLSLALN KSIDPTVAMT GELTLTGKVL RIGGLREKAV AAKRSGAKTI IFPKDNLNDW1080
1081 EELPDNVKEG LEPLAADWYN DIFQKLFKDV NTKEGNSVWK AEFEILDAKK EKD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |

|
MIS1 PIM1 |
| View Details | Gavin AC, et al. (2002) |
|
IDH2 PIM1 SAM1 |
| View Details | Ho Y, et al. (2002) |

|
ARP7 AVO1 BEM3 CCT2 COR1 CYS4 DIG1 DIG2 DOG1 ENP1 FET4 FPR3 GFA1 GUS1 HAS1 HXT6 KSS1 MAM33 MDN1 MKT1 MSE1 NAP1 NPA3 PEX6 PHO84 PIM1 PMA1 PRP8 PYC1 QCR2 RGC1 RPA135 RPN10 RPN6 RVB1 SAP185 SAP190 SEN1 SIT4 SNP1 SRP1 STE11 STE12 STE7 TEC1 TSC11 UBI4 USA1 VAC14 YDR239C YHR033W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
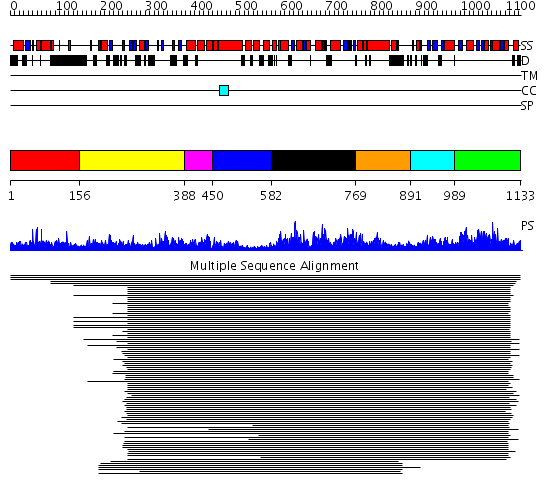
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [156..387] | 71.744727 | ATP-dependent protease La (LON) domain No confident structure predictions are available. | |
| 3 | View Details | [388..449] | 7.54 | delta subunit; delta subunit of DNA polymerase III, N-domain | |
| 4 | View Details | [450..581] | 7.54 | delta subunit; delta subunit of DNA polymerase III, N-domain | |
| 5 | View Details | [582..768] | 62.221849 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 6 | View Details | [769..890] | 62.221849 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 7 | View Details | [891..988] | 2.234996 | View MSA. Confident ab initio structure predictions are available. | |
| 8 | View Details | [989..1133] | 27.136997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)