













| General Information: |
|
| Name(s) found: |
TOR1 /
YJR066W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extrinsic to internal side of plasma membrane
[IDA Golgi membrane [IDA endosome membrane [IDA vacuolar membrane [IDA TORC1 complex [IPI plasma membrane [IDA |
| Biological Process: |
fungal-type cell wall organization
[IMP regulation of cell growth [IMP signal transduction [TAS G1 phase of mitotic cell cycle [IMP ribosome biogenesis [IMP mitochondria-nucleus signaling pathway [IMP meiosis [IMP |
| Molecular Function: |
protein binding
[IPI protein kinase activity [IMP 1-phosphatidylinositol-3-kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPHEEQIWK SKLLKAANND MDMDRNVPLA PNLNVNMNMK MNASRNGDEF GLTSSRFDGV 60
61 VIGSNGDVNF KPILEKIFRE LTSDYKEERK LASISLFDLL VSLEHELSIE EFQAVSNDIN 120
121 NKILELVHTK KTSTRVGAVL SIDTLISFYA YTERLPNETS RLAGYLRGLI PSNDVEVMRL 180
181 AAKTLGKLAV PGGTYTSDFV EFEIKSCLEW LTASTEKNSF SSSKPDHAKH AALLIITALA 240
241 ENCPYLLYQY LNSILDNIWR ALRDPHLVIR IDASITLAKC LSTLRNRDPQ LTSQWVQRLA 300
301 TSCEYGFQVN TLECIHASLL VYKEILFLKD PFLNQVFDQM CLNCIAYENH KAKMIREKIY 360
361 QIVPLLASFN PQLFAGKYLH QIMDNYLEIL TNAPANKIPH LKDDKPQILI SIGDIAYEVG 420
421 PDIAPYVKQI LDYIEHDLQT KFKFRKKFEN EIFYCIGRLA VPLGPVLGKL LNRNILDLMF 480
481 KCPLSDYMQE TFQILTERIP SLGPKINDEL LNLVCSTLSG TPFIQPGSPM EIPSFSRERA 540
541 REWRNKNILQ KTGESNDDNN DIKIIIQAFR MLKNIKSRFS LVEFVRIVAL SYIEHTDPRV 600
601 RKLAALTSCE IYVKDNICKQ TSLHSLNTVS EVLSKLLAIT IADPLQDIRL EVLKNLNPCF 660
661 DPQLAQPDNL RLLFTALHDE SFNIQSVAME LVGRLSSVNP AYVIPSIRKI LLELLTKLKF 720
721 STSSREKEET ASLLCTLIRS SKDVAKPYIE PLLNVLLPKF QDTSSTVAST ALRTIGELSV 780
781 VGGEDMKIYL KDLFPLIIKT FQDQSNSFKR EAALKALGQL AASSGYVIDP LLDYPELLGI 840
841 LVNILKTENS QNIRRQTVTL IGILGAIDPY RQKEREVTST TDISTEQNAP PIDIALLMQG 900
901 MSPSNDEYYT TVVIHCLLKI LKDPSLSSYH TAVIQAIMHI FQTLGLKCVS FLDQIIPTIL 960
961 DVMRTCSQSL LEFYFQQLCS LIIIVRQHIR PHVDSIFQAI KDFSSVAKLQ ITLVSVIEAI1020
1021 SKALEGEFKR LVPLTLTLFL VILENDKSSD KVLSRRVLRL LESFGPNLEG YSHLITPKIV1080
1081 QMAEFTSGNL QRSAIITIGK LAKDVDLFEM SSRIVHSLLR VLSSTTSDEL SKVIMNTLSL1140
1141 LLIQMGTSFA IFIPVINEVL MKKHIQHTIY DDLTNRILNN DVLPTKILEA NTTDYKPAEQ1200
1201 MEAADAGVAK LPINQSVLKS AWNSSQQRTK EDWQEWSKRL SIQLLKESPS HALRACSNLA1260
1261 SMYYPLAKEL FNTAFACVWT ELYSQYQEDL IGSLCIALSS PLNPPEIHQT LLNLVEFMEH1320
1321 DDKALPIPTQ SLGEYAERCH AYAKALHYKE IKFIKEPENS TIESLISINN QLNQTDAAIG1380
1381 ILKHAQQHHS LQLKETWFEK LERWEDALHA YNEREKAGDT SVSVTLGKMR SLHALGEWEQ1440
1441 LSQLAARKWK VSKLQTKKLI APLAAGAAWG LGEWDMLEQY ISVMKPKSPD KEFFDAILYL1500
1501 HKNDYDNASK HILNARDLLV TEISALINES YNRAYSVIVR TQIITEFEEI IKYKQLPPNS1560
1561 EKKLHYQNLW TKRLLGCQKN VDLWQRVLRV RSLVIKPKQD LQIWIKFANL CRKSGRMRLA1620
1621 NKALNMLLEG GNDPSLPNTF KAPPPVVYAQ LKYIWATGAY KEALNHLIGF TSRLAHDLGL1680
1681 DPNNMIAQSV KLSSASTAPY VEEYTKLLAR CFLKQGEWRI ATQPNWRNTN PDAILGSYLL1740
1741 ATHFDKNWYK AWHNWALANF EVISMVQEET KLNGGKNDDD DDTAVNNDNV RIDGSILGSG1800
1801 SLTINGNRYP LELIQRHVVP AIKGFFHSIS LLETSCLQDT LRLLTLLFNF GGIKEVSQAM1860
1861 YEGFNLMKIE NWLEVLPQLI SRIHQPDPTV SNSLLSLLSD LGKAHPQALV YPLTVAIKSE1920
1921 SVSRQKAALS IIEKIRIHSP VLVNQAELVS HELIRVAVLW HELWYEGLED ASRQFFVEHN1980
1981 IEKMFSTLEP LHKHLGNEPQ TLSEVSFQKS FGRDLNDAYE WLNNYKKSKD INNLNQAWDI2040
2041 YYNVFRKITR QIPQLQTLDL QHVSPQLLAT HDLELAVPGT YFPGKPTIRI AKFEPLFSVI2100
2101 SSKQRPRKFS IKGSDGKDYK YVLKGHEDIR QDSLVMQLFG LVNTLLKNDS ECFKRHLDIQ2160
2161 QYPAIPLSPK SGLLGWVPNS DTFHVLIREH RDAKKIPLNI EHWVMLQMAP DYENLTLLQK2220
2221 IEVFTYALDN TKGQDLYKIL WLKSRSSETW LERRTTYTRS LAVMSMTGYI LGLGDRHPSN2280
2281 LMLDRITGKV IHIDFGDCFE AAILREKYPE KVPFRLTRML TYAMEVSGIE GSFRITCENV2340
2341 MRVLRDNKES LMAILEAFAL DPLIHWGFDL PPQKLTEQTG IPLPLINPSE LLRKGAITVE2400
2401 EAANMEAEQQ NETKNARAML VLRRITDKLT GNDIKRFNEL DVPEQVDKLI QQATSIERLC2460
2461 QHYIGWCPFW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
TOR1 VPS52 VPS53 VPS54 |
| View Details | Gavin AC, et al. (2006) |
|
TBF1 TOR1 VID22 |
| View Details | Qiu et al. (2008) |
|
KOG1 LST8 TCO89 TOR1 TOR2 |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
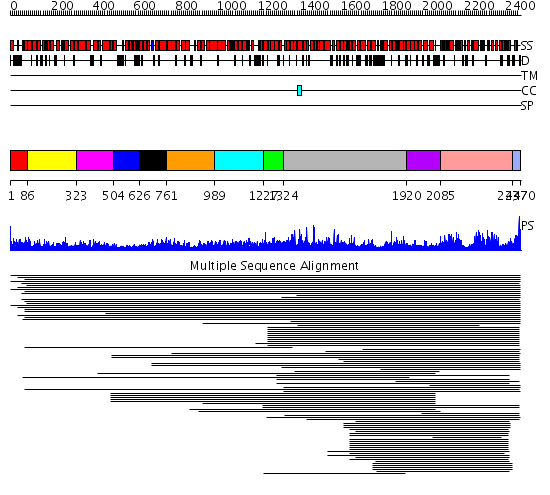
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..322] | 81.221849 | Importin beta | |
| 3 | View Details | [323..503] | 81.221849 | Importin beta | |
| 4 | View Details | [504..625] | 81.221849 | Importin beta | |
| 5 | View Details | [626..760] | 81.221849 | Importin beta | |
| 6 | View Details | [761..988] | 81.221849 | Importin beta | |
| 7 | View Details | [989..1226] | 81.154902 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 8 | View Details | [1227..1323] | N/A | Confident ab initio structure predictions are available. | |
| 9 | View Details | [1324..1919] | 235.376751 | FAT domain No confident structure predictions are available. | |
| 10 | View Details | [1920..2084] | 332.30103 | FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP) | |
| 11 | View Details | [2085..2436] | 123.60206 | Phosphatidylinositol 3- and 4-kinase No confident structure predictions are available. | |
| 12 | View Details | [2437..2470] | 17.130768 | FATC domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)