













| General Information: |
|
| Name(s) found: |
TOR2 /
YKL203C
[SGD]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2473 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TORC2 complex
[IPI mitochondrion [IDA extrinsic to internal side of plasma membrane [IDA vacuolar membrane [IDA TORC1 complex [IPI plasma membrane [IDA |
| Biological Process: |
cytoskeleton organization
[IMP regulation of cell growth [TAS signal transduction [TAS G1 phase of mitotic cell cycle [TAS ribosome biogenesis [IMP Rho protein signal transduction [IMP endocytosis [IMP actin filament reorganization involved in cell cycle [TAS establishment or maintenance of actin cytoskeleton polarity [IMP |
| Molecular Function: |
protein binding
[IPI protein serine/threonine kinase activity [IDA 1-phosphatidylinositol-3-kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKYINKYTT PPNLLSLRQR AEGKHRTRKK LTHKSHSHDD EMSTTSNTDS NHNGPNDSGR 60
61 VITGSAGHIG KISFVDSELD TTFSTLNLIF DKLKSDVPQE RASGANELST TLTSLAREVS 120
121 AEQFQRFSNS LNNKIFELIH GFTSSEKIGG ILAVDTLISF YLSTEELPNQ TSRLANYLRV 180
181 LIPSSDIEVM RLAANTLGRL TVPGGTLTSD FVEFEVRTCI DWLTLTADNN SSSSKLEYRR 240
241 HAALLIIKAL ADNSPYLLYP YVNSILDNIW VPLRDAKLII RLDAAVALGK CLTIIQDRDP 300
301 ALGKQWFQRL FQGCTHGLSL NTNDSVHATL LVFRELLSLK APYLRDKYDD IYKSTMKYKE 360
361 YKFDVIRREV YAILPLLAAF DPAIFTKKYL DRIMVHYLRY LKNIDMNAAN NSDKPFILVS 420
421 IGDIAFEVGS SISPYMTLIL DNIREGLRTK FKVRKQFEKD LFYCIGKLAC ALGPAFAKHL 480
481 NKDLLNLMLN CPMSDHMQET LMILNEKIPS LESTVNSRIL NLLSISLSGE KFIQSNQYDF 540
541 NNQFSIEKAR KSRNQSFMKK TGESNDDITD AQILIQCFKM LQLIHHQYSL TEFVRLITIS 600
601 YIEHEDSSVR KLAALTSCDL FIKDDICKQT SVHALHSVSE VLSKLLMIAI TDPVAEIRLE 660
661 ILQHLGSNFD PQLAQPDNLR LLFMALNDEI FGIQLEAIKI IGRLSSVNPA YVVPSLRKTL 720
721 LELLTQLKFS NMPKKKEESA TLLCTLINSS DEVAKPYIDP ILDVILPKCQ DASSAVASTA 780
781 LKVLGELSVV GGKEMTRYLK ELMPLIINTF QDQSNSFKRD AALTTLGQLA ASSGYVVGPL 840
841 LDYPELLGIL INILKTENNP HIRRGTVRLI GILGALDPYK HREIEVTSNS KSSVEQNAPS 900
901 IDIALLMQGV SPSNDEYYPT VVIHNLMKIL NDPSLSIHHT AAIQAIMHIF QNLGLRCVSF 960
961 LDQIIPGIIL VMRSCPPSQL DFYFQQLGSL ISIVKQHIRP HVEKIYGVIR EFFPIIKLQI1020
1021 TIISVIESIS KALEGEFKRF VPETLTFFLD ILENDQSNKR IVPIRILKSL VTFGPNLEDY1080
1081 SHLIMPIVVR MTEYSAGSLK KISIITLGRL AKNINLSEMS SRIVQALVRI LNNGDRELTK1140
1141 ATMNTLSLLL LQLGTDFVVF VPVINKALLR NRIQHSVYDQ LVNKLLNNEC LPTNIIFDKE1200
1201 NEVPERKNYE DEMQVTKLPV NQNILKNAWY CSQQKTKEDW QEWIRRLSIQ LLKESPSACL1260
1261 RSCSSLVSVY YPLARELFNA SFSSCWVELQ TSYQEDLIQA LCKALSSSEN PPEIYQMLLN1320
1321 LVEFMEHDDK PLPIPIHTLG KYAQKCHAFA KALHYKEVEF LEEPKNSTIE ALISINNQLH1380
1381 QTDSAIGILK HAQQHNELQL KETWYEKLQR WEDALAAYNE KEAAGEDSVE VMMGKLRSLY1440
1441 ALGEWEELSK LASEKWGTAK PEVKKAMAPL AAAAWGLEQW DEIAQYTSVM KSQSPDKEFY1500
1501 DAILCLHRNN FKKAEVHIFN ARDLLVTELS ALVNESYNRA YNVVVRAQII AELEEIIKYK1560
1561 KLPQNSDKRL TMRETWNTRL LGCQKNIDVW QRILRVRSLV IKPKEDAQVR IKFANLCRKS1620
1621 GRMALAKKVL NTLLEETDDP DHPNTAKASP PVVYAQLKYL WATGLQDEAL KQLINFTSRM1680
1681 AHDLGLDPNN MIAQSVPQQS KRVPRHVEDY TKLLARCFLK QGEWRVCLQP KWRLSNPDSI1740
1741 LGSYLLATHF DNTWYKAWHN WALANFEVIS MLTSVSKKKQ EGSDASSVTD INEFDNGMIG1800
1801 VNTFDAKEVH YSSNLIHRHV IPAIKGFFHS ISLSESSSLQ DALRLLTLWF TFGGIPEATQ1860
1861 AMHEGFNLIQ IGTWLEVLPQ LISRIHQPNQ IVSRSLLSLL SDLGKAHPQA LVYPLMVAIK1920
1921 SESLSRQKAA LSIIEKMRIH SPVLVDQAEL VSHELIRMAV LWHEQWYEGL DDASRQFFGE1980
1981 HNTEKMFAAL EPLYEMLKRG PETLREISFQ NSFGRDLNDA YEWLMNYKKS KDVSNLNQAW2040
2041 DIYYNVFRKI GKQLPQLQTL ELQHVSPKLL SAHDLELAVP GTRASGGKPI VKISKFEPVF2100
2101 SVISSKQRPR KFCIKGSDGK DYKYVLKGHE DIRQDSLVMQ LFGLVNTLLQ NDAECFRRHL2160
2161 DIQQYPAIPL SPKSGLLGWV PNSDTFHVLI REHREAKKIP LNIEHWVMLQ MAPDYDNLTL2220
2221 LQKVEVFTYA LNNTEGQDLY KVLWLKSRSS ETWLERRTTY TRSLAVMSMT GYILGLGDRH2280
2281 PSNLMLDRIT GKVIHIDFGD CFEAAILREK FPEKVPFRLT RMLTYAMEVS GIEGSFRITC2340
2341 ENVMKVLRDN KGSLMAILEA FAFDPLINWG FDLPTKKIEE ETGIQLPVMN ANELLSNGAI2400
2401 TEEEVQRVEN EHKNAIRNAR AMLVLKRITD KLTGNDIRRF NDLDVPEQVD KLIQQATSVE2460
2461 NLCQHYIGWC PFW |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
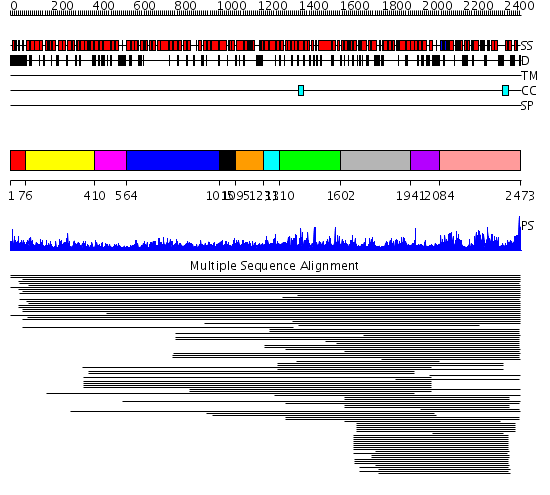
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..409] | 11.19 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [410..563] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [564..1014] | 21.428996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1015..1094] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [1095..1230] | 2.195995 | View MSA. Confident ab initio structure predictions are available. | |
| 7 | View Details | [1231..1309] | N/A | Confident ab initio structure predictions are available. | |
| 8 | View Details | [1310..1601] | 13.097997 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1602..1940] | 1.092973 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1941..2083] | 347.218487 | FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP) | |
| 11 | View Details | [2084..2473] | 13.116964 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)