













| General Information: |
|
| Name(s) found: |
PYC1 /
YGL062W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1178 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
gluconeogenesis
[IMP NADPH regeneration [TAS] |
| Molecular Function: |
pyruvate carboxylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQRKFAGLR DNFNLLGEKN KILVANRGEI PIRIFRTAHE LSMQTVAIYS HEDRLSTHKQ 60
61 KADEAYVIGE VGQYTPVGAY LAIDEIISIA QKHQVDFIHP GYGFLSENSE FADKVVKAGI 120
121 TWIGPPAEVI DSVGDKVSAR NLAAKANVPT VPGTPGPIET VEEALDFVNE YGYPVIIKAA 180
181 FGGGGRGMRV VREGDDVADA FQRATSEART AFGNGTCFVE RFLDKPKHIE VQLLADNHGN 240
241 VVHLFERDCS VQRRHQKVVE VAPAKTLPRE VRDAILTDAV KLAKECGYRN AGTAEFLVDN 300
301 QNRHYFIEIN PRIQVEHTIT EEITGIDIVA AQIQIAAGAS LPQLGLFQDK ITTRGFAIQC 360
361 RITTEDPAKN FQPDTGRIEV YRSAGGNGVR LDGGNAYAGT IISPHYDSML VKCSCSGSTY 420
421 EIVRRKMIRA LIEFRIRGVK TNIPFLLTLL TNPVFIEGTY WTTFIDDTPQ LFQMVSSQNR 480
481 AQKLLHYLAD VAVNGSSIKG QIGLPKLKSN PSVPHLHDAQ GNVINVTKSA PPSGWRQVLL 540
541 EKGPAEFARQ VRQFNGTLLM DTTWRDAHQS LLATRVRTHD LATIAPTTAH ALAGRFALEC 600
601 WGGATFDVAM RFLHEDPWER LRKLRSLVPN IPFQMLLRGA NGVAYSSLPD NAIDHFVKQA 660
661 KDNGVDIFRV FDALNDLEQL KVGVDAVKKA GGVVEATVCF SGDMLQPGKK YNLDYYLEIA 720
721 EKIVQMGTHI LGIKDMAGTM KPAAAKLLIG SLRAKYPDLP IHVHTHDSAG TAVASMTACA 780
781 LAGADVVDVA INSMSGLTSQ PSINALLASL EGNIDTGINV EHVRELDAYW AEMRLLYSCF 840
841 EADLKGPDPE VYQHEIPGGQ LTNLLFQAQQ LGLGEQWAET KRAYREANYL LGDIVKVTPT 900
901 SKVVGDLAQF MVSNKLTSDD VRRLANSLDF PDSVMDFFEG LIGQPYGGFP EPFRSDVLRN 960
961 KRRKLTCRPG LELEPFDLEK IREDLQNRFG DVDECDVASY NMYPRVYEDF QKMRETYGDL1020
1021 SVLPTRSFLS PLETDEEIEV VIEQGKTLII KLQAVGDLNK KTGEREVYFD LNGEMRKIRV1080
1081 ADRSQKVETV TKSKADMHDP LHIGAPMAGV IVEVKVHKGS LIKKGQPVAV LSAMKMEMII1140
1141 SSPSDGQVKE VFVSDGENVD SSDLLVLLED QVPVETKA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PYC1 PYC2 |
| View Details | Krogan NJ, et al. (2006) |
|
PYC1 PYC2 SPS1 TPM1 |
| View Details | Ho Y, et al. (2002) |
|
ACC1 ACH1 ADH2 ADH3 ADR1 ARP7 CDC12 CDC60 DEF1 DIG1 DIG2 DOG1 DOG2 ECM10 FAA1 FET4 GCD11 GDE1 GFA1 GND1 GUS1 HAS1 HEF3 HIR3 HXT6 IDH2 IMD4 ISA2 KGD1 KSS1 MDH1 MET18 MET6 MKT1 MSE1 MSK1 NAP1 NGL2 PHO84 PIM1 PMA1 PRO2 PTC6 PYC1 PYC2 RAD1 RET1 RGC1 RGD1 RPA135 RPA190 RPA49 RPB5 RPC19 RPC25 RPC34 RPC40 RPC82 RPN10 RPN3 RPO26 RPO31 RVS167 SEC27 SEC53 SEN1 SML1 STE12 STE7 TBS1 TEC1 TFP1 TIF1, TIF2 TMA19 TPS1 TSA2 UBI4 VMA22 YDR239C YFL042C YHR033W YHR112C |
| View Details | Qiu et al. (2008) |

|
PYC1 PYC2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
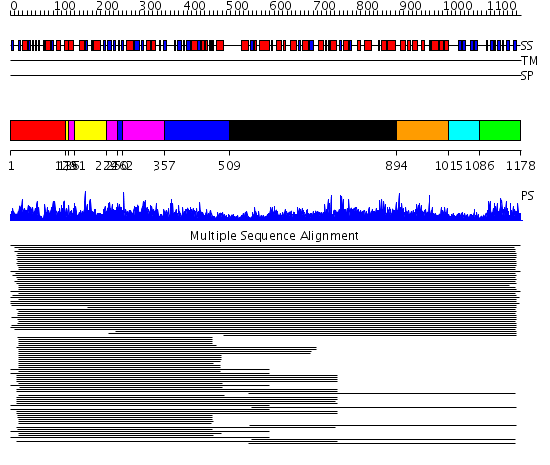
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 1115.218487 | No description for 1lsrA_ was found. | |
| 2 | View Details | [129..135] [151..223] |
1115.218487 | No description for 1lsrA_ was found. | |
| 3 | View Details | [136..150] [224..249] [262..356] |
1115.218487 | No description for 1lsrA_ was found. | |
| 4 | View Details | [250..261] [357..508] |
1115.218487 | No description for 1lsrA_ was found. | |
| 5 | View Details | [509..893] | 9.06 | Betaine-homocysteine S-methyltransferase | |
| 6 | View Details | [894..1014] | 9.41 | Dihydrodipicolinate synthase | |
| 7 | View Details | [1015..1085] | N/A | Confident ab initio structure predictions are available. | |
| 8 | View Details | [1086..1178] | 81.927757 | Biotin carboxyl carrier domain of transcarboxylase (TC 1.3S) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)