













| General Information: |
|
| Name(s) found: |
KGD1 /
YIL125W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1014 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[TAS mitochondrial oxoglutarate dehydrogenase complex [IDA mitochondrion [IDA mitochondrial nucleoid [IDA |
| Biological Process: |
tricarboxylic acid cycle
[TAS 2-oxoglutarate metabolic process [TAS |
| Molecular Function: |
oxoglutarate dehydrogenase (succinyl-transferring) activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRFVSSQTC RYSSRGLLKT SLLKNASTVK IVGRGLATTG TDNFLSTSNA TYIDEMYQAW 60
61 QKDPSSVHVS WDAYFKNMSN PKIPATKAFQ APPSISNFPQ GTEAAPLGTA MTGSVDENVS 120
121 IHLKVQLLCR AYQVRGHLKA HIDPLGISFG SNKNNPVPPE LTLDYYGFSK HDLDKEINLG 180
181 PGILPRFARD GKSKMSLKEI VDHLEKLYCS SYGVQYTHIP SKQKCDWLRE RIEIPEPYQY 240
241 TVDQKRQILD RLTWATSFES FLSTKFPNDK RFGLEGLESV VPGIKTLVDR SVELGVEDIV 300
301 LGMAHRGRLN VLSNVVRKPN ESIFSEFKGS SARDDIEGSG DVKYHLGMNY QRPTTSGKYV 360
361 NLSLVANPSH LESQDPVVLG RTRALLHAKN DLKEKTKALG VLLHGDAAFA GQGVVYETMG 420
421 FLTLPEYSTG GTIHVITNNQ IGFTTDPRFA RSTPYPSDLA KAIDAPIFHV NANDVEAVTF 480
481 IFNLAAEWRH KFHTDAIIDV VGWRKHGHNE TDQPSFTQPL MYKKIAKQKS VIDVYTEKLI 540
541 SEGTFSKKDI DEHKKWVWNL FEDAFEKAKD YVPSQREWLT AAWEGFKSPK ELATEILPHE 600
601 PTNVPESTLK ELGKVLSSWP EGFEVHKNLK RILKNRGKSI ETGEGIDWAT GEALAFGTLV 660
661 LDGQNVRVSG EDVERGTFSQ RHAVLHDQQS EAIYTPLSTL NNEKADFTIA NSSLSEYGVM 720
721 GFEYGYSLTS PDYLVMWEAQ FGDFANTAQV IIDQFIAGGE QKWKQRSGLV LSLPHGYDGQ 780
781 GPEHSSGRLE RFLQLANEDP RYFPSEEKLQ RQHQDCNFQV VYPTTPANLF HILRRQQHRQ 840
841 FRKPLALFFS KQLLRHPLAR SSLSEFTEGG FQWIIEDIEH GKSIGTKEET KRLVLLSGQV 900
901 YTALHKRRES LGDKTTAFLK IEQLHPFPFA QLRDSLNSYP NLEEIVWCQE EPLNMGSWAY 960
961 TEPRLHTTLK ETDKYKDFKV RYCGRNPSGA VAAGSKSLHL AEEDAFLKDV FQQS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
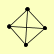
|
KGD1 KGD2 LPD1 YMR31 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
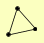
|
KGD1 KGD2 LPD1 |
| View Details | Krogan NJ, et al. (2006) |
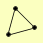
|
KGD1 KGD2 STE5 |
| View Details | Gavin AC, et al. (2006) |
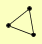
|
KGD1 KGD2 YMR31 |
| View Details | Gavin AC, et al. (2002) |
|
CLU1 KAP123 KGD1 KGD2 LEA1 LPD1 SAM1 YMR31 |
| View Details | Ho Y, et al. (2002) |

|
ACO1 ATG12 CAP1 CAP2 CAR1 CAR2 CDC15 CDC33 CDC60 CIC1 COF1 COR1 CPA2 CPR1 CPR3 CRM1 CYS4 DBP8 DEF1 DNL4 DUG2 DUT1 EMP24 FAA1 FET3 GDE1 GND1 GPH1 GSP1 GSP2 GUS1 HEF3 HSP104 HTA1 KAP122 KGD1 KRE6 MAK16 MDH1 MET10 MET18 NAN1 NOP15 NOP2 NOP7 NSA1 OYE2 PFK1 PGM2 PRO2 PTC6 PYC1 RAD1 RNA1 RNR1 RNR2 RPF2 RPN1 RPN12 RPN5 RPN6 RPN8 RPN9 RPT1 RPT2 RRT6 SCS2 SEC26 SEC53 TEF4 TEM1 THI22 TIF1, TIF2 TIF6 TMA29 TPS1 TPS2 UTP10 UTP9 YDL086W YFR016C YGR066C YRB1 YTM1 |
| View Details | Qiu et al. (2008) |
|
ALD4 ARG5,6 CIT1 IDH1 IDP1 KGD1 KGD2 LPD1 MDH1 PDA1 PDB1 SDH4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample zds1 from august 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
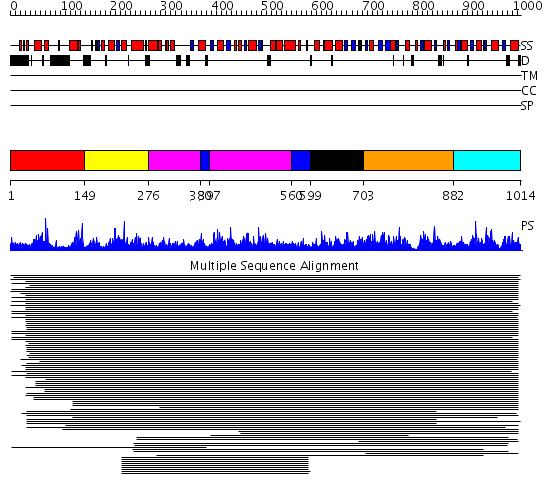
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 2.055989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [149..275] | 1.297993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [276..379] [397..559] |
70.154902 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain | |
| 4 | View Details | [380..396] [560..598] |
70.154902 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain | |
| 5 | View Details | [599..702] | 70.154902 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain | |
| 6 | View Details | [703..881] | 75.0 | E1-beta subunit of pyruvate dehydrogenase, Pyr module; E1-beta subunit of pyruvate dehydrogenase, C-domain | |
| 7 | View Details | [882..1014] | 75.0 | E1-beta subunit of pyruvate dehydrogenase, Pyr module; E1-beta subunit of pyruvate dehydrogenase, C-domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)