













| General Information: |
|
| Name(s) found: |
KRE6 /
YPR159W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IDA Golgi cis cisterna [IDA |
| Biological Process: |
cellular cell wall organization
[IMP 1,6-beta-glucan biosynthetic process [IMP |
| Molecular Function: |
glucosidase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLRNLTETH NFSSTNLDTD GTGDDHDGAP LSSSPSFGQQ NDNSTNDNAG LTNPFMGSDE 60
61 ESNARDGESL SSSVHYQPQG SDSSLLHDNS RLDLSQNKGV SDYKGYYSRN NSRAVSTAND 120
121 NSFLQPPHRA IASSPSLNSN LSKNDILSPP EFDRYPLVGS RVTSMTQLNH HGRSPTSSPG 180
181 NESSASFSSN PFLGEQDFSP FGGYPASSFP LMIDEKEEDD YLHNPDPEEE ARLDRRRFID 240
241 DFKYMDKRSA SGLAGVLLLF LAAIFIFIVL PALTFTGAID HESNTEEVTY LTQYQYPQLS 300
301 AIRTSLVDPD TPDTAKTREA MDGSKWELVF SDEFNAEGRT FYDGDDPYWT APDVHYDATK 360
361 DLEWYSPDAS TTVNGTLQLR MDAFKNHGLY YRSGMLQSWN KVCFTQGALE ISANLPNYGR 420
421 VSGLWPGLWT MGNLGRPGYL ASTQGVWPYS YESCDAGITP NQSSPDGISY LPGQKLSICT 480
481 CDGEDHPNQG VGRGAPEIDV LEGETDTKIG VGIASQSLQI APFDIWYMPD YDFIEVYNFT 540
541 TTTMNTYAGG PFQQAVSAVS TLNVTWYEFG EYGGYFQKYA IEYLNDDDNG YIRWFVGDTP 600
601 TYTIHAKALH PDGNIGWRRI SKEPMSIILN LGISNNWAYI DWQYIFFPVV MSIDYVRIYQ 660
661 PSNAISVTCD PSDYPTYDYI QSHLNAFQNA NLTTWEDAGY TFPKNILTGK CTSSKFKLSS 720
721 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Lockshon D, et al. (2006) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
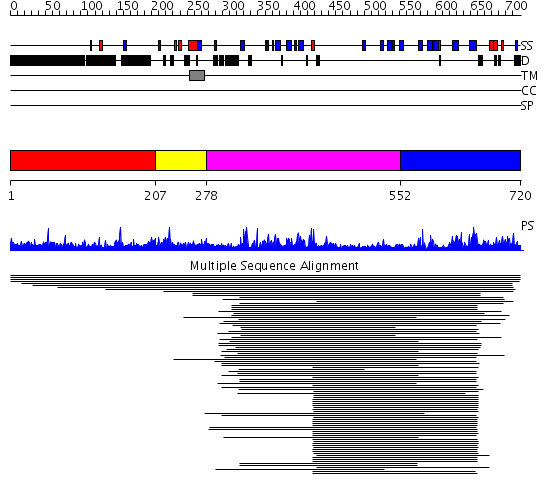
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [207..277] | 14.886057 | Beta-glucan synthesis-associated protein (SKN1) No confident structure predictions are available. | |
| 3 | View Details | [278..551] | 14.09691 | kappa-Carrageenase, catalytic | |
| 4 | View Details | [552..720] | 32.0 | Bacillus 1-3,1-4-beta-glucanase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|