













| General Information: |
|
| Name(s) found: |
TPS2 /
YDR074W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 896 amino acids |
Gene Ontology: |
|
| Cellular Component: |
alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)
[IGI mitochondrion [IDA |
| Biological Process: |
trehalose biosynthetic process
[IMP response to stress [IDA |
| Molecular Function: |
trehalose-phosphatase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTTAQDNSP KKRQRIINCV TQLPYKIQLG ESNDDWKISA TTGNSALFSS LEYLQFDSTE 60
61 YEQHVVGWTG EITRTERNLF TREAKEKPQD LDDDPLYLTK EQINGLTTTL QDHMKSDKEA 120
121 KTDTTQTAPV TNNVHPVWLL RKNQSRWRNY AEKVIWPTFH YILNPSNEGE QEKNWWYDYV 180
181 KFNEAYAQKI GEVYRKGDII WIHDYYLLLL PQLLRMKFND ESIIIGYFHH APWPSNEYFR 240
241 CLPRRKQILD GLVGANRICF QNESFSRHFV SSCKRLLDAT AKKSKNSSNS DQYQVSVYGG 300
301 DVLVDSLPIG VNTTQILKDA FTKDIDSKVL SIKQAYQNKK IIIGRDRLDS VRGVVQKLRA 360
361 FETFLAMYPE WRDQVVLIQV SSPTANRNSP QTIRLEQQVN ELVNSINSEY GNLNFSPVQH 420
421 YYMRIPKDVY LSLLRVADLC LITSVRDGMN TTALEYVTVK SHMSNFLCYG NPLILSEFSG 480
481 SSNVLKDAIV VNPWDSVAVA KSINMALKLD KEEKSNLESK LWKEVPTIQD WTNKFLSSLK 540
541 EQASSNDDME RKMTPALNRP VLLENYKQAK RRLFLFDYDG TLTPIVKDPA AAIPSARLYT 600
601 ILQKLCADPH NQIWIISGRD QKFLNKWLGG KLPQLGLSAE HGCFMKDVSC QDWVNLTEKV 660
661 DMSWQVRVNE VMEEFTTRTP GSFIERKKVA LTWHYRRTVP ELGEFHAKEL KEKLLSFTDD 720
721 FDLEVMDGKA NIEVRPRFVN KGEIVKRLVW HQHGKPQDML KGISEKLPKD EMPDFVLCLG 780
781 DDFTDEDMFR QLNTIETCWK EKYPDQKNQW GNYGFYPVTV GSASKKTVAK AHLTDPQQVL 840
841 ETLGLLVGDV SLFQSAGTVD LDSRGHVKNS ESSLKSKLAS KAYVMKRSAS YTGAKV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
TPS1 TPS2 TPS3 TSL1 |
| View Details | Krogan NJ, et al. (2006) |
|
TPS1 TPS2 TPS3 TSL1 |
| View Details | Gavin AC, et al. (2002) |
|
CTR3 DAL3 GLN1 GTO3 IML1 MCM2 MKS1 REG1 TIF1, TIF2 TPS1 TPS2 TPS3 TSC10 TSL1 YIL177C |
| View Details | Ho Y, et al. (2002) |
|
GND1 GPH1 HSP104 KGD1 NAN1 PFK1 SCS2 TPS2 UTP10 UTP9 |
| View Details | Qiu et al. (2008) |
|
TPS1 TPS2 TPS3 TSL1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
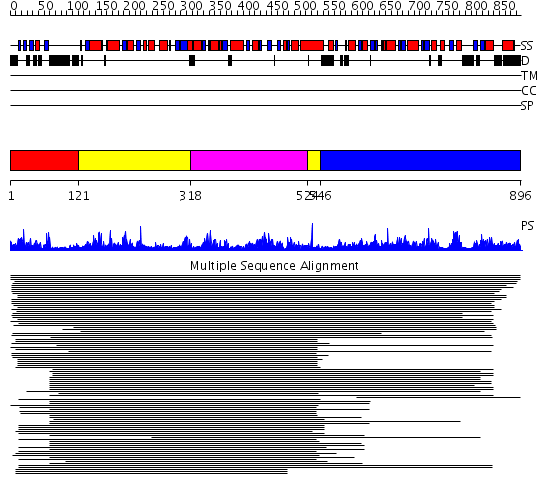
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [121..317] [524..545] |
13.39 | UDP-N-acetylglucosamine 2-epimerase | |
| 3 | View Details | [318..523] | 13.39 | UDP-N-acetylglucosamine 2-epimerase | |
| 4 | View Details | [546..896] | 25.886997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)