













| General Information: |
|
| Name(s) found: |
TPS3 /
YMR261C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1054 amino acids |
Gene Ontology: |
|
| Cellular Component: |
alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)
[IGI |
| Biological Process: |
trehalose biosynthetic process
[IGI response to stress [IMP |
| Molecular Function: |
alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity
[IMP trehalose-phosphatase activity [IGI enzyme regulator activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIIVASLFL PYTPQFEADV TNSDTAKLVE SSMIKVDCNN QELSNNKQER SSSVTSASSH 60
61 YIGLPQEAQI NGEPLQRANV GSPATGVNYH NEMEMLSSEQ FLEELTANAT HAANSGIPPA 120
121 NNPVSSGSTA QRPSVEEFFS APSARVCSPS QEASASSISA SRSSAHHNDL SSSLMKNPNL 180
181 SFDSHPPRVR SSSKSAVITP VSKSVPDVDP AVVDVAKVRE EFQQQASLPS MKRVSGSTAG 240
241 DSSIASSSSN LRYSQQFQDN FIEDTDSEDD IDSDLETDAT KKYNVPKFGG YSNNAKLRAS 300
301 LMRNSYELFK HLPWTIVDSD KGNGSLKNAV NIAVAEKTVK EPVSWVGTMG IPTDELPHEV 360
361 CHKISKKLEQ DFSSFPVVTD DITFKGAYKN YAKQILWPTL HYQIPDNPNS KAFEDHSWDY 420
421 YQKVNQKFSD RIVSVYKPGD TIWIHDYHLM LVPQMVREKL PKAKIGFFLH VSFPSSEVFR 480
481 CLANRERILE GIIGANFVGF QTKEYKRHFL QTCNRLLAAD VSNDEVKYHC NIVSVMYAPI 540
541 GIDYYHLTSQ LRNGSVLEWR QLIKERWRNK KLIVCRDQFD RIRGLQKKML AYERFLIENP 600
601 EYIEKVVLIQ ICIGKSSDPE YERQIMVVVD RINSLSSNIS ISQPVVFLHQ DLDFAQYLAL 660
661 NCEADVFLVD ALREGMNLTC HEFIVSSFEK NAPLLLSEFT GSSSVLKEGA ILINPWDINH 720
721 VAQSIKRSLE MSPEEKRRRW KKLFKSVIEH DSDNWITKCF EYINNAWESN QETSTVFNLA 780
781 PEKFCADYKA SKKHLFIFKI SEPPTSRMLS LLSELSSNNI VYVLSSFTKN TFESLYNGVL 840
841 NIGLIAENGA YVRVNGSWYN IVEELDWMKE VAKIFDEKVE RLPGSYYKIA DSMIRFHTEN 900
901 ADDQDRVPTV IGEAITHINT LFDDRDIHAY VHKDIVFVQQ TGLALAAAEF LMKFYNSGVS 960
961 PTDNSRISLS RTSSSMSVGN NKKHFQNQVD FVCVSGSTSP IIEPLFKLVK QEVEKNNLKF1020
1021 GYTILYGSSR STYAKEHING VNELFTILHD LTAA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
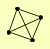
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
TPS1 TPS2 TPS3 TSL1 |
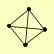
| View Details | Krogan NJ, et al. (2006) |
|
TPS1 TPS2 TPS3 TSL1 |
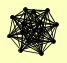
| View Details | Gavin AC, et al. (2002) |
|
CTR3 DAL3 GLN1 GTO3 IML1 MCM2 MKS1 REG1 TIF1, TIF2 TPS1 TPS2 TPS3 TSC10 TSL1 YIL177C |
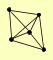
| View Details | Qiu et al. (2008) |
|
GIP2 REG1 TPS1 TPS2 TPS3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
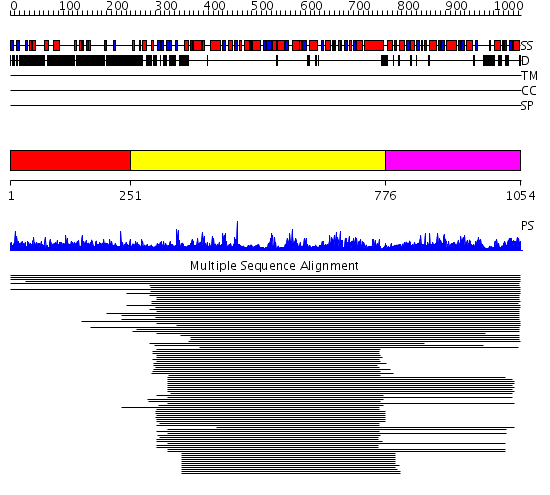
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..250] | 48.290997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [251..775] | 254.356547 | Glycosyltransferase family 20 No confident structure predictions are available. | |
| 3 | View Details | [776..1054] | 71.920819 | Trehalose-phosphatase No confident structure predictions are available. | |
| 4 | View Details | [779..1054] | 11.09691 | Crystal structure of trehalose-6-phosphate phosphatase related protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)