













| General Information: |
|
| Name(s) found: |
MCM2 /
YBL023C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 868 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA pre-replicative complex [IDA cytoplasm [IDA MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[TAS pre-replicative complex assembly [IPI DNA unwinding involved in replication [TAS |
| Molecular Function: |
chromatin binding
[TAS ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDNRRRRRE EDDSDSENEL PPSSPQQHFR GGMNPVSSPI GSPDMINPEG DDNEVDDVPD 60
61 IDEVEEQMNE VDLMDDNMYE DYAADHNRDR YDPDQVDDRE QQELSLSERR RIDAQLNERD 120
121 RLLRNVAYID DEDEEQEGAA QLDEMGLPVQ RRRRRRQYED LENSDDDLLS DMDIDPLREE 180
181 LTLESLSNVK ANSYSEWITQ PNVSRTIARE LKSFLLEYTD ETGRSVYGAR IRTLGEMNSE 240
241 SLEVNYRHLA ESKAILALFL AKCPEEMLKI FDLVAMEATE LHYPDYARIH SEIHVRISDF 300
301 PTIYSLRELR ESNLSSLVRV TGVVTRRTGV FPQLKYVKFN CLKCGSILGP FFQDSNEEIR 360
361 ISFCTNCKSK GPFRVNGEKT VYRNYQRVTL QEAPGTVPPG RLPRHREVIL LADLVDVSKP 420
421 GEEVEVTGIY KNNYDGNLNA KNGFPVFATI IEANSIKRRE GNTANEGEEG LDVFSWTEEE 480
481 EREFRKISRD RGIIDKIISS MAPSIYGHRD IKTAVACSLF GGVPKNVNGK HSIRGDINVL 540
541 LLGDPGTAKS QILKYVEKTA HRAVFATGQG ASAVGLTASV RKDPITKEWT LEGGALVLAD 600
601 KGVCLIDEFD KMNDQDRTSI HEAMEQQSIS ISKAGIVTTL QARCSIIAAA NPNGGRYNST 660
661 LPLAQNVSLT EPILSRFDIL CVVRDLVDEE ADERLATFVV DSHVRSHPEN DEDREGEELK 720
721 NNGESAIEQG EDEINEQLNA RQRRLQRQRK KEEEISPIPQ ELLMKYIHYA RTKIYPKLHQ 780
781 MDMDKVSRVY ADLRRESIST GSFPITVRHL ESILRIAESF AKMRLSEFVS SYDLDRAIKV 840
841 VVDSFVDAQK VSVRRQLRRS FAIYTLGH |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
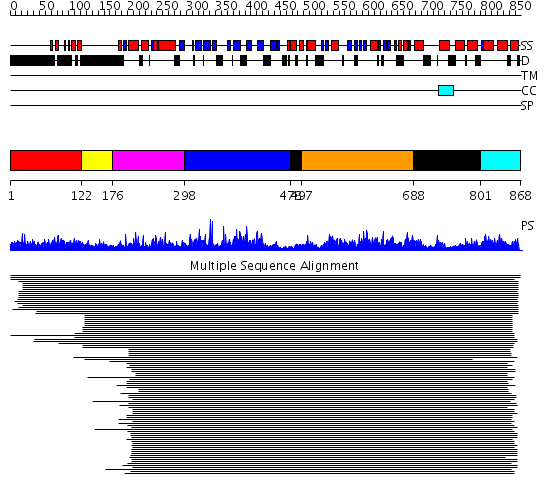
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 1.015984 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [122..175] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [176..297] | 1.095989 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [298..477] | 7.221849 | MCM2/3/5 family No confident structure predictions are available. | |
| 5 | View Details | [478..496] [688..800] |
2.522879 | Replication factor C | |
| 6 | View Details | [497..687] | 2.522879 | Replication factor C | |
| 7 | View Details | [801..868] | 1.494987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)