













| General Information: |
|
| Name(s) found: |
NMD3 /
YHR170W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 518 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
ribosomal large subunit export from nucleus
[IMP ribosome biogenesis [RCA ribosomal large subunit assembly [IMP |
| Molecular Function: |
RNA binding
[ISS protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEFTPIDPHQ HQNAATLLCC NCGTPIDGST GLVMCYDCIK LTVDITQGIP REANISFCRN 60
61 CERFLQPPGQ WIRAELESRE LLAICLRRLK GLTKVRLVDA SFIWTEPHSR RIRIKLTVQG 120
121 EAMTNTIIQQ TFEVEYIVIA MQCPDCARSY TTNTWRATVQ IRQKVPHKRT FLFLEQLILK 180
181 HNAHVDTISI SEAKDGLDFF YAQKNHAVKM IDFLNAVVPI KHKKSEELIS QDTHTGASTY 240
241 KFSYSVEIVP ICKDDLVVLP KKLAKSMGNI SQFVLCSKIS NTVQFMDPTT LQTADLSPSV 300
301 YWRAPFNALA DVTQLVEFIV LDVDSTGISR GNRVLADITV ARTSDLGVND QVYYVRSHLG 360
361 GICHAGDSVM GYFIANSNYN SDLFDGLNID YVPDVVLVKK LYQRKSKKSR HWKLKRMAKE 420
421 HKDIDASLDY NSRAQKQEME RAEKDYELFL QELEEDAELR QSVNLYKNRE ANVPPEEHEM 480
481 DEDEDEDAPQ INIDELLDEL DEMTLEDGVE NTPVESQQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
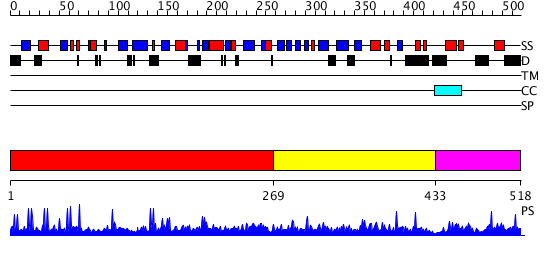
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..268] | 10.023955 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [269..432] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [433..518] | 1.009994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)