













| General Information: |
|
| Name(s) found: |
MCM3 /
YEL032W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 971 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA pre-replicative complex [IDA cytoplasm [IDA MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[TAS pre-replicative complex assembly [IPI DNA unwinding involved in replication [TAS |
| Molecular Function: |
chromatin binding
[TAS ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGSTGFDGD ATTFFAPDAV FGDRVRRFQE FLDTFTSYRD SVRSIQVYNS NNAANYNDDQ 60
61 DDADERDLLG DDDGDDLEKE KKAASSTSLN ILPHRIIISL DDLREFDRSF WSGILVEPAY 120
121 FIPPAEKALT DLADSMDDVP HPNASAVSSR HPWKLSFKGS FGAHALSPRT LTAQHLNKLV 180
181 SVEGIVTKTS LVRPKLIRSV HYAAKTGRFH YRDYTDATTT LTTRIPTPAI YPTEDTEGNK 240
241 LTTEYGYSTF IDHQRITVQE MPEMAPAGQL PRSIDVILDD DLVDKTKPGD RVNVVGVFKS 300
301 LGAGGMNQSN SNTLIGFKTL ILGNTVYPLH ARSTGVAARQ MLTDFDIRNI NKLSKKKDIF 360
361 DILSQSLAPS IYGHDHIKKA ILLMLMGGVE KNLENGSHLR GDINILMVGD PSTAKSQLLR 420
421 FVLNTASLAI ATTGRGSSGV GLTAAVTTDR ETGERRLEAG AMVLADRGVV CIDEFDKMTD 480
481 VDRVAIHEVM EQQTVTIAKA GIHTTLNARC SVIAAANPVF GQYDVNRDPH QNIALPDSLL 540
541 SRFDLLFVVT DDINEIRDRS ISEHVLRTHR YLPPGYLEGE PVRERLNLSL AVGEDADINP 600
601 EEHSNSGAGV ENEGEDDEDH VFEKFNPLLQ AGAKLAKNKG NYNGTEIPKL VTIPFLRKYV 660
661 QYAKERVIPQ LTQEAINVIV KNYTDLRNDD NTKKSPITAR TLETLIRLAT AHAKVRLSKT 720
721 VNKVDAKVAA NLLRFALLGE DIGNDIDEEE SEYEEALSKR SPQKSPKKRQ RVRQPASNSG 780
781 SPIKSTPRRS TASSVNATPS SARRILRFQD DEQNAGEDDN DIMSPLPADE EAELQRRLQL 840
841 GLRVSPRRRE HLHAPEEGSS GPLTEVGTPR LPNVSSAGQD DEQQQSVISF DNVEPGTIST 900
901 GRLSLISGII ARLMQTEIFE EESYPVASLF ERINEELPEE EKFSAQEYLA GLKIMSDRNN 960
961 LMVADDKVWR V |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CDC45 CDC6 DPB11 DPB2 DPB3 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 SLD3 |
| View Details | Ho Y, et al. (2002) |

|
BMS1 CDC53 CIC1 COF1 CTF4 DBP10 DED81 DIA2 ENP1 FPR3 ILS1 KRE33 KRI1 LST4 MCM2 MCM3 MCM5 NIP7 NMD3 NOP12 RRP12 SEH1 SKP1 SLT2 SPB1 SSF1 SSF2 SSZ1 TIF6 URB1 UTP10 YAK1 YBL104C |
| View Details | Qiu et al. (2008) |
|
CDC45 CDC6 DPB11 DPB2 DPB3 DPB4 HCS1 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 PCC1 POB3 POL1 POL2 POL3 POL31 POL32 RFC1 RFC4 SLD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
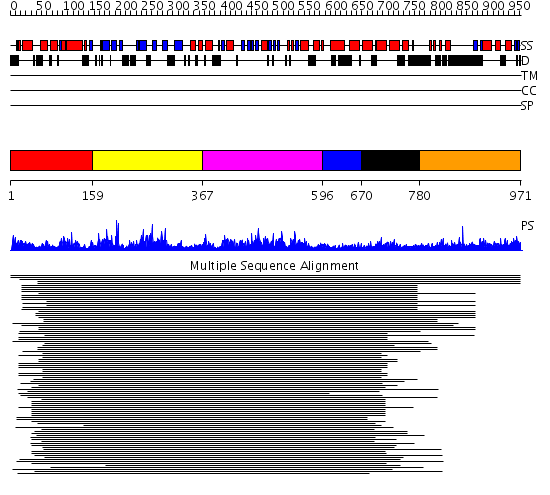
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 1.098987 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [159..366] | 9.086186 | MCM2/3/5 family No confident structure predictions are available. | |
| 3 | View Details | [367..595] | 40.69897 | ATPase subunit of magnesium chelatase, BchI | |
| 4 | View Details | [596..669] | 40.69897 | ATPase subunit of magnesium chelatase, BchI | |
| 5 | View Details | [670..779] | 7.44 | ATPase subunit of magnesium chelatase, BchI | |
| 6 | View Details | [780..971] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)