













| General Information: |
|
| Name(s) found: |
CDC53 /
YDL132W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 815 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SCF ubiquitin ligase complex
[IDA |
| Biological Process: |
G2/M transition of mitotic cell cycle
[IGI G1/S transition of mitotic cell cycle [IMP SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [IDA protein ubiquitination during ubiquitin-dependent protein catabolic process [IDA |
| Molecular Function: |
DNA replication origin binding
[IPI ubiquitin-protein ligase activity [IDA protein binding, bridging [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSETLPRSDD LEATWNFIEP GINQILGNEK NQASTSKRVY KILSPTMYME VYTAIYNYCV 60
61 NKSRSSGHFS TDSRTGQSTI LVGSEIYEKL KNYLKNYILN FKQSNSETFL QFYVKRWKRF 120
121 TIGAIFLNHA FDYMNRYWVQ KERSDGKRHI FDVNTLCLMT WKEVMFDPSK DVLINELLDQ 180
181 VTLGREGQII QRSNISTAIK SLVALGIDPQ DLKKLNLNVY IQVFEKPFLK KTQEYYTQYT 240
241 NDYLEKHSVT EYIFEAHEII KREEKAMTIY WDDHTKKPLS MALNKVLITD HIEKLENEFV 300
301 VLLDARDIEK ITSLYALIRR DFTLIPRMAS VFENYVKKTG ENEISSLLAM HKHNIMKNEN 360
361 ANPKKLALMT AHSLSPKDYI KKLLEVHDIF SKIFNESFPD DIPLAKALDN ACGAFININE 420
421 FALPAGSPKS ATSKTSEMLA KYSDILLKKA TKPEVASDMS DEDIITIFKY LTDKDAFETH 480
481 YRRLFAKRLI HGTSTSAEDE ENIIQRLQAA NSMEYTGKIT KMFQDIRLSK ILEDDFAVAL 540
541 KNEPDYSKAK YPDLQPFVLA ENMWPFSYQE VEFKLPKELV PSHEKLKESY SQKHNGRILK 600
601 WLWPLCRGEL KADIGKPGRM PFNFTVTLFQ MAILLLYNDA DVLTLENIQE GTSLTIQHIA 660
661 AAMVPFIKFK LIQQVPPGLD ALVKPETQFK LSRPYKALKT NINFASGVKN DILQSLSGGG 720
721 HDNHGNKLGN KRLTEDERIE KELNTERQIF LEACIVRIMK AKRNLPHTTL VNECIAQSHQ 780
781 RFNAKVSMVK RAIDSLIQKG YLQRGDDGES YAYLA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
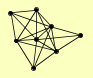
|
CDC34 CDC53 HRT1 MET30 RTG2 RTT101 SAF1 SKP1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
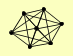
|
CDC34 CDC4 CDC53 GRR1 MET30 SGT1 SKP1 |
| View Details | Krogan NJ, et al. (2006) |
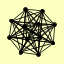
|
CAK1 CDC34 CDC53 CDC9 CRT10 GRR1 MEP3 MET30 MMS1 POR1 RTT101 SAF1 |
| View Details | Ho Y, et al. (2002) |

|
AAH1 ADH2 ADR1 BBC1 BMS1 BOP2 CDC39 CDC4 CDC53 CIC1 COF1 CPR1 CRM1 CTF4 DAS1 DBP10 DED81 DIA2 DUR1,2 ECM29 ECM33 ENP1 FAA4 FOL2 FPR3 GAL3 GCN1 GRR1 GUF1 HRT1 HRT3 HTB2 HYP2 IDH1 ILS1 KRE33 KRI1 LST4 MCM2 MCM3 MCM5 MDN1 MET30 MET31 MET4 MKT1 MMS1 MYO2 NIP7 NMD3 NOP12 PDC5 PDC6 PFK1 PGM2 PMA2 POR1 PRB1 PTC1 RIX7 RPA190 RPN1 RPN8 RRP12 RTT101 RUB1 SAF1 SAH1 SEC27 SEH1 SGT1 SIS1 SKP1 SLT2 SPB1 SRV2 SSF1 SSF2 SSZ1 STI1 TEF4 THI21 TIF6 TPS1 UBI4 UFO1 URB1 UTP10 VPS13 YAK1 YAR009C YBL104C YBR139W YDR131C YDR306C YGP1 YLR035C-A YLR352W |
| View Details | Qiu et al. (2008) |
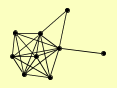
|
APC9 CDC34 CDC4 CDC53 DIA2 HRT1 MET30 SKP1 UFO1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
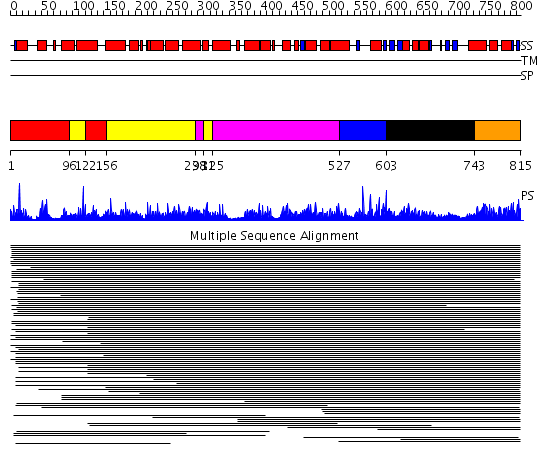
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] [122..155] |
2110.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 2 | View Details | [96..121] [156..297] [311..324] |
2110.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 3 | View Details | [298..310] [325..526] |
2110.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 4 | View Details | [527..602] | 2110.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 5 | View Details | [603..742] | 2110.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 6 | View Details | [743..815] | 2110.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)