













| General Information: |
|
| Name(s) found: |
DBP10 /
YDL031W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 995 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[TAS nucleolus [IDA |
| Biological Process: |
ribosome biogenesis
[RCA ribosomal large subunit assembly [IMP |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGVQKRKRD LEDQDDNGSE EDDIAFDIAN EIALNDSESD ANDSDSEVEA DYGPNDVQDV 60
61 IEYSSDEEEG VNNKKKAENK DIKKKKNSKK EIAAFPMLEM SDDENNASGK TQTGDDEDDV 120
121 NEYFSTNNLE KTKHKKGSFP SFGLSKIVLN NIKRKGFRQP TPIQRKTIPL ILQSRDIVGM 180
181 ARTGSGKTAA FILPMVEKLK SHSGKIGARA VILSPSRELA MQTFNVFKDF ARGTELRSVL 240
241 LTGGDSLEEQ FGMMMTNPDV IIATPGRFLH LKVEMNLDLK SVEYVVFDEA DRLFEMGFQE 300
301 QLNELLASLP TTRQTLLFSA TLPNSLVDFV KAGLVNPVLV RLDAETKVSE NLEMLFLSSK 360
361 NADREANLLY ILQEIIKIPL ATSEQLQKLQ NSNNEADSDS DDENDRQKKR RNFKKEKFRK 420
421 QKMPAANELP SEKATILFVP TRHHVEYISQ LLRDCGYLIS YIYGTLDQHA RKRQLYNFRA 480
481 GLTSILVVTD VAARGVDIPM LANVINYTLP GSSKIFVHRV GRTARAGNKG WAYSIVAENE 540
541 LPYLLDLELF LGKKILLTPM YDSLVDVMKK RWIDEGKPEY QFQPPKLSYT KRLVLGSCPR 600
601 LDVEGLGDLY KNLMSSNFDL QLAKKTAMKA EKLYYRTRTS ASPESLKRSK EIISSGWDAQ 660
661 NAFFGKNEEK EKLDFLAKLQ NRRNKETVFE FTRNPDDEMA VFMKRRRKQL APIQRKATER 720
721 RELLEKERMA GLHILSKMKF WKGDDVETGY TVSEDALKEF EDAHQLLEAQ ENENKKKKKP 780
781 KSFKDPTFFL SHYAPAGDIQ DKQLQITNGF ANDAAQAAYD LNSDDKVQVH KQTATVKWDK 840
841 KRKKYVNTQG IDNKKYIIGE SGQKIAASFR SGRFDDWSKA RNLKPLKVGS RETSIPSNLL 900
901 EDPSQGPAAN GRTVRGKFKH KQMKAPKMPD KHRDNYYSQK KKVEKALQSG ISVKGYNNAP 960
961 GLRSELKSTE QIRKDRIIAE KKRAKNARPS KKRKF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
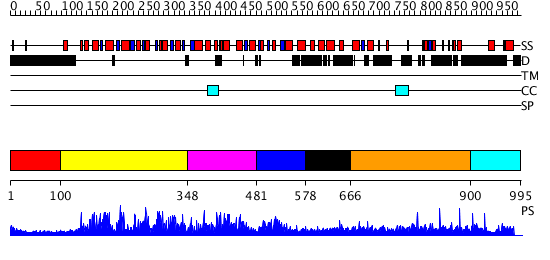
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..347] | 419.9794 | Putative DEAD box RNA helicase | |
| 3 | View Details | [348..480] | 419.9794 | Putative DEAD box RNA helicase | |
| 4 | View Details | [481..577] | 13.84 | No description for 1gkuB was found. | |
| 5 | View Details | [578..665] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [666..899] | 1.014962 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [900..995] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)