













| General Information: |
|
| Name(s) found: |
RPG1 /
YBR079C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 964 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA cytoplasm [IDA ribosome [TAS eukaryotic translation initiation factor 3 complex [IDA |
| Biological Process: |
translational initiation
[IDA |
| Molecular Function: |
translation initiation factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPPPFRPEN AIKRADELIS VGEKQAALQS LHDFITARRI RWATPSTVEP VVFKFLEIGV 60
61 ELKKGKLLKD GLHQYKKLIQ GSTEGLVSVG AVARKFIDLV ESKIASEQTR ADELQKQEID 120
121 DDLEGGVTPE NLLISVYESD QSVAGFNDEA ITSWLRFTWE SYRAVLDLLR NNALLEITYS 180
181 GVVKKTMHFC LKYQRKNEFK RLAEMLRQHL DAANYQQSKS GNNLVDLSDA DTLQRYLDQR 240
241 FQQVDVSVKL ELWHEAYRSI EDVFHLMKIS KRAPKPSTLA NYYENLVKVF FVSGDPLLHT 300
301 TAWKKFYKLY STNPRATEEE FKTYSSTIFL SAISTQLDEI PSIGYDPHLR MYRLLNLDAK 360
361 PTRKEMLQSI IEDESIYGKV DEELKELYDI IEVNFDVDTV KQQLENLLVK LSSKTYFSQY 420
421 IAPLRDVIMR RVFVAASQKF TTVSQSELYK LATLPAPLDL SAWDIEKSLL QAAVEDYVSI 480
481 TIDHESAKVT FAKDPFDIFA STASKEVSEE ENTEPEVQEE KEETDEALGP QETEDGEEKE 540
541 EESDPVIIRN SYIHNKLLEL SNVLHDVDSF NNASYMEKVR IARETLIKKN KDDLEKISKI 600
601 VDERVKRSQE QKQKHMEHAA LHAEQDAEVR QQRILEEKAA IEAKLEEEAH RRLIEKKKRE 660
661 FEAIKEREIT KMITEVNAKG HVYIDPNEAK SLDLDTIKQV IIAEVSKNKS ELESRMEYAM 720
721 KKLDHTERAL RKVELPLLQK EVDKLQETDT ANYEAMKKKI VDAAKAEYEA RMADRKNLVM 780
781 VYDDYLKFKE HVSGTKESEL AAIRNQKKAE LEAAKKARIE EVRKRRYEEA IARRKEEIAN 840
841 AERQKRAQEL AEATRKQREI EEAAAKKSTP YSFRAGNREP PSTPSTLPKA TVSPDKAKLD 900
901 MIAQKQREME EAIEQRLAGR TAGGSSPATP ATPATPATPT PSSGPKKMTM AEKLRAKRLA 960
961 KGGR |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
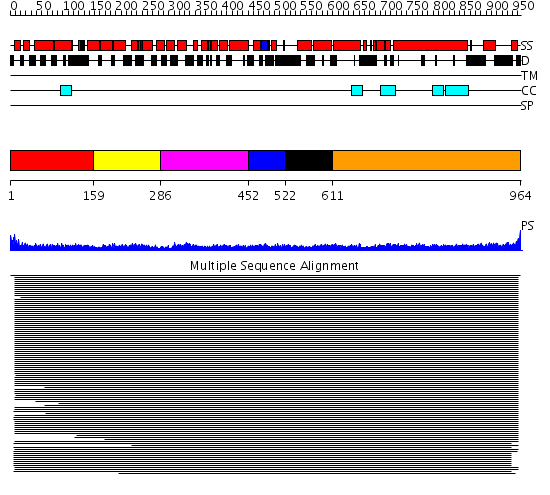
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 11.47 | Transcription factor MalT domain III | |
| 2 | View Details | [159..285] | 11.47 | Transcription factor MalT domain III | |
| 3 | View Details | [286..451] | 11.47 | Transcription factor MalT domain III | |
| 4 | View Details | [452..521] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [522..610] | 8.69897 | Tropomyosin | |
| 6 | View Details | [611..964] | 13.0 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)