













| General Information: |
|
| Name(s) found: |
HIF1 /
YLL022C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 385 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA histone acetyltransferase complex [IDA |
| Biological Process: |
histone acetylation
[IDA chromatin silencing at telomere [IGI nucleosome assembly [IDA |
| Molecular Function: |
histone binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLRAEDVLA NGTSRHKVQI DMERQVQIAK DLLAQKKFLE AAKRCQQTLD SLPKDGLLPD 60
61 PELFTIFAQA VYNMEVQNSG NLFGDALLAG DDGSGSESES EPESDVSNGE EGNENGQTEI 120
121 PNSRMFQFDQ EEEDLTGDVD SGDSEDSGEG SEEEEENVEK EEERLALHEL ANFSPANEHD 180
181 DEIEDVSQLR KSGFHIYFEN DLYENALDLL AQALMLLGRP TADGQSLTEN SRLRIGDVYI 240
241 LMGDIEREAE MFSRAIHHYL KALGYYKTLK PAEQVTEKVI QAEFLVCDAL RWVDQVPAKD 300
301 KLKRFKHAKA LLEKHMTTRP KDSELQQARL AQIQDDIDEV QENQQHGSKR PLSQPTTSIG 360
361 FPALEKPLGD FNDLSQLVKK KPRRH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
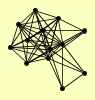
|
ASF1 HAT1 HAT2 HHF1, HHF2 HHT1, HHT2 HIF1 MAM33 ORC2 POB3 PSH1 PTC4 SPT16 YJR154W |
| View Details | Krogan NJ, et al. (2006) |
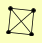
|
HIF1 SAP4 SPT2 TRI1 |
| View Details | Gavin AC, et al. (2006) |
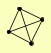
|
HAT1 HAT2 HHF1, HHF2 HIF1 |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |
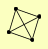
|
HAT1 HAT2 HIF1 RTN1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
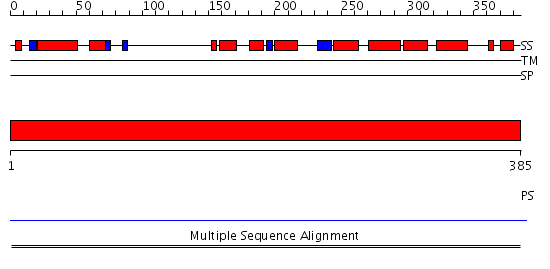
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..385] | 7.63 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)