













| General Information: |
|
| Name(s) found: |
BRX1 /
YOL077C
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 291 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IPI |
| Biological Process: |
ribosome biogenesis
[RCA ribosomal large subunit assembly [IPI |
| Molecular Function: |
5S rRNA binding
[IPI rRNA primary transcript binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSIYKALAG KSKDNKSEKK QGNVKQFMNK QRTLLISSRG VNYRHRHLIQ DLSGLLPHSR 60
61 KEPKLDTKKD LQQLNEIAEL YNCNNVLFFE ARKHQDLYLW LSKPPNGPTI KFYIQNLHTM 120
121 DELNFTGNCL KGSRPVLSFD QRFESSPHYQ LIKELLVHNF CVPPNARKSK PFIDHVMSFS 180
181 IVDDKIWVRT YEISHSTKNK EEYEDGEEDI SLVEIGPRFV MTVILILEGS FGGPKIYENK 240
241 QYVSPNVVRA QIKQQAAEEA KSRAEAAVER KIKRRENVLA ADPLSNDALF K |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
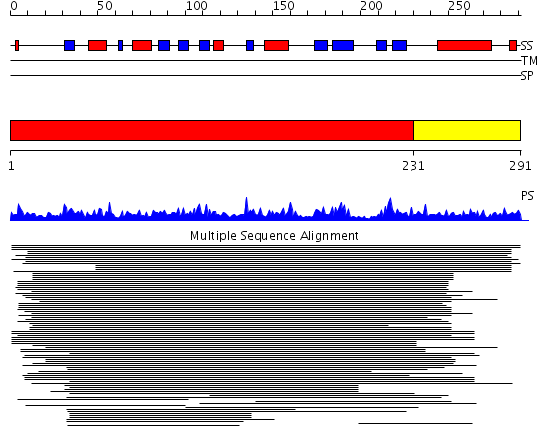
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..230] | 9.060986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [231..291] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)