













| General Information: |
|
| Name(s) found: |
LTV1 /
YKL143W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 463 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA late endosome membrane [IDA cytoplasm [IDA preribosome, small subunit precursor [IDA |
| Biological Process: |
ribosomal small subunit export from nucleus
[IGI response to oxidative stress [IMP ribosomal small subunit biogenesis [IMP response to osmotic stress [IMP ribosome biogenesis [RCA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKKFSSKNS QRYVVVHRPH DDPSFYDTDA SAHVLVPVSN PNKTSPEADL RKKDVSSTKP 60
61 KGRRAHVGEA ALYGINFDDS EYDYTQHLKP IGLDPENSIF IASKGNEQKV EKKNIEDLFI 120
121 EPKYRRDEIE KDDALPVFQR GMAKPEYLLH QQDTTDEIRG FKPDMNPALR EVLEALEDEA 180
181 YVVNDDVVVE DISKKTQLQG DNYGEEEKED DIFAQLLGSG EAKDEDEFED EFDEWDIDNV 240
241 ENFEDENYVK EMAQFDNIEN LEDLENIDYQ ADVRRFQKDN SILEKHNSDD EFSNAGLDSV 300
301 NPSEEEDVLG ELPSIQDKSK TGKKKRKSRQ KKGAMSDVSG FSMSSSAIAR TETMTVLDDQ 360
361 YDQIINGYEN YEEELEEDEE QNYQPFDMSA ERSDFESMLD DFLDNYELES GGRKLAKKDK 420
421 EIERLKEAAD EVSKGKLSQR RNRERQEKKK LEKVTNTLSS LKF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
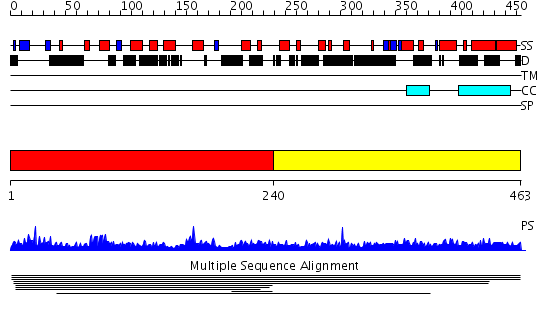
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..239] | 3.007954 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [240..463] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)