













| General Information: |
|
| Name(s) found: |
MDS3 /
YGL197W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1487 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
negative regulation of sporulation resulting in formation of a cellular spore
[IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLLQPSTCF CYPLKLPPLP LTSDSNEFDE CARKRLTLDY RTGSAVTLTR SNIFVHGGLT 60
61 IPLNLPVVNS MQLQKELILF FAKEKNNGSS FRNLNEWISK ETFFLDLMSR TWYRVKTSFD 120
121 QRTEELLKAE SSSAKADNDT NEIRTDIKKG KSLESPLKER LFHSLCYLDG CLYIFGGLTV 180
181 SPQSGYELIA TNELWKLDLN TKKWSLLSDD PQIARRFNHT MHVKNENNDN RDTKLIIVGG 240
241 LNNMDQPVKK IDIYNISQNC WHPKTIPKQP MEITTNVNGI PLALSKDQNF SILVENNEAN 300
301 VPALAFYMRS DQIDEYLGKD SSKIKENSPI VALPLLSESQ GIRMPSNPAL PKKLLNVPYE 360
361 LLAPTGDYFG FNIIIGGFHP NYQSSNFHCF IYDINSGKWS RVRTACPDCD INKHRFWRVF 420
421 VWKSHHQTIL LGTKTDDYYS PSVQRFDHLS TFGLPLVNIF NKTIQLPHHK ISASSLPIPI 480
481 ENFAKHKDTP LKKVSFTSSA TSQFENYIRY IAPPLEMSSI QSVFPPYAMV LGKDALEIYG 540
541 KPLSDFEFIT SEGDSIGIPC YLLRKRWGRY FDMLLSQSYT KVCADYETTD TQSTLIKFSP 600
601 HSSRNSSKAV RQEGRLSSSG SLDNYFEKNF PIFARTSVSE AQNTQPQVAN ADAKAPNTPS 660
661 TSDEPSSSSS SDLYSTPHYQ RNNDEEDDED PVSPKPVSKS NSIYRPIRKT ESSSTTSSSN 720
721 GMIFRVPFKE KAAVTSNTEA LLESNLSLQE LSRRRSSLMS IPSGELLRSS ISEAEHQRRA 780
781 SHPLTSSPLF EDSGTPCGKQ LQQLQQHTIQ NPHNHLSPRR FSRSARSSIS YVSSSSDRRG 840
841 NSISSRSTSD SFGTPPVLGV LNVPLPPQTR EPNEPPPPCP AMSTGSNTRR SNTLTDYMHS 900
901 NKASPFSSRR SSHIGRRSST PETENAFSAT PRASLDGQML GKSLKEGSTS QYTQPRMNSF 960
961 PKANETIQTP TSSNNEWSRQ SVTSNTDSFD SLQSNFALEL EPLLTPRSLY MPWPTSTVRA1020
1021 FAEFFYTGQV NSKWLLAPVA LDLLVMAKIY EIPLLYKLIL EVLYSILAKK EESLSLICTS1080
1081 LMETFRTKTL NSYKGDEEKT NTYLTSNDNY QELLKLKVSL ENIDNGYYDP DLLRKQSRAQ1140
1141 SSSTQESSGS ANGEKTATGA GSLETSSTNV PTVFAGGPRD SHNSVGSIGF PNSMNIQGSR1200
1201 RSTSGFSPRV KMKSSLSKEI DPKTFYEEYE PKEGKSFDDN DDQQTNIGSF NLHLFDMNYG1260
1261 SISSSSTNSI SSSDLEEKEE QEQLQDLLEI EREDSAEILD ARFRNKEDDK VTKDISNDKK1320
1321 RNYLPHEKNN LKAKEGKETR DVREEEEEFD FGLGMLSLNK IKREAKHVDK VDDSVDPLFK1380
1381 SSAFPQSPIR AYGSTTRTSS ASGKPFRDNR SFNAFSVLTL ENMASANALP PVDYVIKSIY1440
1441 RTTVLVNDIR LMVRCMDCIE LSKNLRALKK KTMEDISKLK GISKPSP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC25 MDS3 SAP185 YBR225W |
| View Details | Krogan NJ, et al. (2006) |
|
CDC25 MDS3 YBR225W |
| View Details | Gavin AC, et al. (2006) |
|
CDC25 HRK1 MDS3 SAP185 SIT4 |
| View Details | Gavin AC, et al. (2002) |
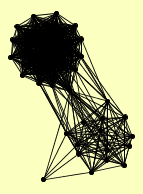
|
ACC1 AVT4 BEM2 BLM10 CCT2 CDC25 DAM1 DBP9 ECM29 EST3 FAB1 GFA1 HRK1 INO4 KAP95 LYS12 MDS3 MIA40 MRPL3 NUD1 PDA1 PDB1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE8 PRE9 PSE1 PUP3 RGR1 RPT3 RPT5 RRP12 SAP155 SAP185 SAP190 SCL1 SIT4 SLX9 SPA2 SRP1 SWT21 ULP1 YBR225W YFL006W YMR310C YRA1 |
| View Details | Ho Y, et al. (2002) |

|
ABP1 ACO1 AIM41 ATP4 BUD14 CAR2 CDC25 CKA2 COR1 CRZ1 CYS4 DCP1 DCP2 DNM1 EDC3 EDE1 EGD2 ENP1 FPR3 GAS1 GCN3 GLC7 GPH1 HHF1, HHF2 HRR25 HSP104 HXT7 HYP2 IPP1 LOC1 LSP1 LTV1 MDH1 MDS3 MGE1 NSA1 NUG1 OYE2 PEX19 PIL1 PIN4 PTC4 PUF3 RIO2 RPC19 RPM2 SAP185 SAP190 SAS10 SEC2 SEC23 SES1 SFB3 SGM1 SIT4 TGL1 TMA19 TSR1 VMA4 YBR225W YER138C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
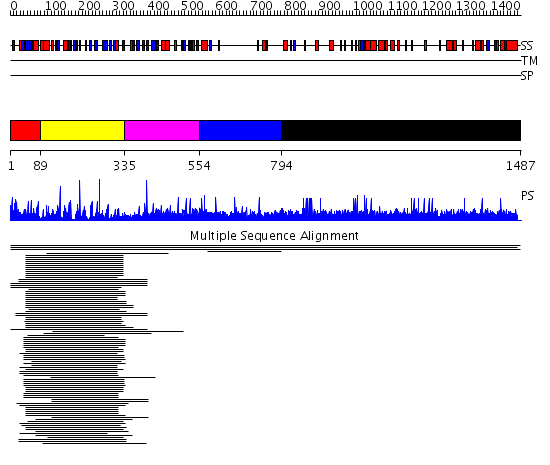
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..334] | 4.522879 | Galactose oxidase, C-terminal domain; Galactose oxidase, N-terminal domain; Galactose oxidase, central domain | |
| 3 | View Details | [335..553] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [554..793] | 1.003934 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [794..1487] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [794..884] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [885..959] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [960..1194] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1195..1326] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1327..1487] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)