













| General Information: |
|
| Name(s) found: |
CDC25 /
YLR310C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA plasma membrane [IDA |
| Biological Process: |
traversing start control point of mitotic cell cycle
[IMP Ras protein signal transduction [TAS replicative cell aging [IMP |
| Molecular Function: |
Ras guanyl-nucleotide exchange factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDTNTSIPN TSSAREAGNA SQTPSISSSS NTSTTTNTES SSASLSSSPS TSELTSIRPI 60
61 GIVVAAYDFN YPIKKDSSSQ LLSVQQGETI YILNKNSSGW WDGLVIDDSN GKVNRGWFPQ 120
121 NFGRPLRDSH LRKHSHPMKK YSSSKSSRRS SLNSLGNSAY LHVPRNPSKS RRGSSTLSAS 180
181 LSNAHNAETS SGHNNTVSMN NSPFSAPNDA SHITPQSSNF NSNASLSQDM TKSADGSSEM 240
241 NTNAIMNNNE TNLQTSGEKA GPPLVAEETI KILPLEEIEM IINGIRSNIA STWSPIPLIT 300
301 KTSDYKLVYY NKDLDIYCSE LPLISNSIME SDDICDSEPK FPPNDHLVNL YTRDLRKNAN 360
361 IEDSSTRSKQ SESEQNRSSL LMEKQDSKET DGNNNSINDD DNNNENNKNE FNEAGPSSLN 420
421 SLSAPDLTQN IQSRVVAPSR SSILAKSDIF YHYSRDIKLW TELQDLTVYY TKTAHKMFLK 480
481 ENRLNFTKYF DLISDSIVFT QLGCRLMQHE IKAKSCSKEI KKIFKGLISS LSRISINSHL 540
541 YFDSAFHRKK MDTMNDKDND NQENNCSRTE GDDGKIEVDS VHDLVSVPLS GKRNVSTSTT 600
601 DTLTPMRSSF STVNENDMEN FSVLGPRNSV NSVVTPRTSI QNSTLEDFSP SNKNFKSAKS 660
661 IYEMVDVEFS KFLRHVQLLY FVLQSSVFSD DNTLPQLLPR FFKGSFSGGS WTNPFSTFIT 720
721 DEFGNATKNK AVTSNEVTAS SSKNSSISRI PPKMADAIAS ASGYSANSET NSQIDLKASS 780
781 AASGSVFTPF NRPSHNRTFS RARVSKRKKK YPLTVDTLNT MKKKSSQIFE KLNNATGEHL 840
841 KIISKPKSRI RNLEINSSTY EQINQNVLLL EILENLDLSI FINLKNLIKT PSILLDLESE 900
901 EFLVHAMSSV SSVLTEFFDI KQAFHDIVIR LIMTTQQTTL DDPYLFSSMR SNFPVGHHEP 960
961 FKNISNTPLV KGPFHKKNEQ LALSLFHVLV SQDVEFNNLE FLNNSDDFKD ACEKYVEISN1020
1021 LACIIVDQLI EERENLLNYA ARMMKNNLTA ELLKGEQEKW FDIYSEDYSD DDSENDEAII1080
1081 DDELGSEDYI ERKAANIEKN LPWFLTSDYE TSLVYDSRGK IRGGTKEALI EHLTSHELVD1140
1141 AAFNVTMLIT FRSILTTREF FYALIYRYNL YPPEGLSYDD YNIWIEKKSN PIKCRVVNIM1200
1201 RTFLTQYWTR NYYEPGIPLI LNFAKMVVSE KIPGAEDLLQ KINEKLINEN EKEPVDPKQQ1260
1261 DSVSAVVQTT KRDNKSPIHM SSSSLPSSAS SAFFRLKKLK LLDIDPYTYA TQLTVLEHDL1320
1321 YLRITMFECL DRAWGTKYCN MGGSPNITKF IANANTLTNF VSHTIVKQAD VKTRSKLTQY1380
1381 FVTVAQHCKE LNNFSSMTAI VSALYSSPIY RLKKTWDLVS TESKDLLKNL NNLMDSKRNF1440
1441 VKYRELLRSV TDVACVPFFG VYLSDLTFTF VGNPDFLHNS TNIINFSKRT KIANIVEEII1500
1501 SFKRFHYKLK RLDDIQTVIE ASLENVPHIE KQYQLSLQVE PRSGNTKGST HASSASGTKT1560
1561 AKFLSEFTDD KNGNFLKLGK KKPPSRLFR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
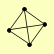
|
CDC25 MDS3 SAP185 YBR225W |
| View Details | Krogan NJ, et al. (2006) |
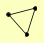
|
CDC25 MDS3 YBR225W |
| View Details | Gavin AC, et al. (2006) |
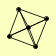
|
CDC25 HRK1 MDS3 SAP185 SIT4 |
| View Details | Gavin AC, et al. (2002) |
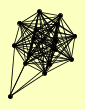
|
ACC1 AVT4 BEM2 CCT2 CDC25 FAB1 HRK1 MDS3 MIA40 MRPL3 SAP155 SAP185 SAP190 SIT4 SWT21 YBR225W |
| View Details | Ho Y, et al. (2002) |
|
CDC25 CKA2 CKI1 DCP2 EDC3 EDE1 ENP1 FPR3 GCR2 GIN4 GLC7 HOM3 HRR25 IRA1 IRA2 MDS3 MYO2 MYO4 NAP1 PMD1 PRS2 PRS3 PRS5 RIM11 RPM2 SEC23 SFB3 SGM1 SIT4 TGL1 TPS1 TSL1 TSR1 YDR170W-A YER138C YER160C YJR027W YJR028W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
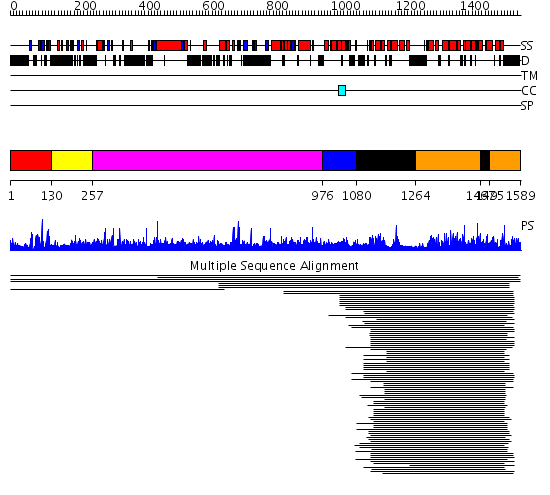
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 47.69897 | c-src protein tyrosine kinase; c-src tyrosine kinase | |
| 2 | View Details | [130..256] | 47.69897 | c-src protein tyrosine kinase; c-src tyrosine kinase | |
| 3 | View Details | [257..975] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [976..1079] | 3.221849 | Son of sevenless-1 (sos-1) | |
| 5 | View Details | [1080..1263] [1467..1494] |
488.0 | Son of sevenless protein homolog 1 (sos-1) | |
| 6 | View Details | [1264..1466] [1495..1589] |
488.0 | Son of sevenless protein homolog 1 (sos-1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)