













| General Information: |
|
| Name(s) found: |
IRA2 /
YOL081W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 3079 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
negative regulation of Ras protein signal transduction
[IMP negative regulation of cAMP biosynthetic process [IMP positive regulation of Ras GTPase activity [IMP response to stress [IMP |
| Molecular Function: |
Ras GTPase activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQPTKNKKK EHGTDSKSSR MTRTLVNHIL FERILPILPV ESNLSTYSEV EEYSSFISCR 60
61 SVLINVTVSR DANAMVEGTL ELIESLLQGH EIISDKGSSD VIESILIILR LLSDALEYNW 120
121 QNQESLHYND ISTHVEHDQE QKYRPKLNSI LPDYSSTHSN GNKHFFHQSK PQALIPELAS 180
181 KLLESCAKLK FNTRTLQILQ NMISHVHGNI LTTLSSSILP RHKSYLTRHN HPSHCKMIDS 240
241 TLGHILRFVA ASNPSEYFEF IRKSVQVPVT QTHTHSHSHS HSLPSSVYNS IVPHFDLFSF 300
301 IYLSKHNFKK YLELIKNLSV TLRKTIYHCL LLHYSAKAIM FWIMARPAEY YELFNLLKDN 360
361 NNEHSKSLNT LNHTLFEEIH STFNVNSMIT TNQNAHQGSS SPSSSSPSSP PSSSSSDNNN 420
421 QNIIAKSLSR QLSHHQSYIQ QQSERKLHSS WTTNSQSSTS LSSSTSNSTT TDFSTHTQPG 480
481 EYDPSLPDTP TMSNITISAS SLLSQTPTPT TQLQQRLNSA AAAAAAAASP SNSTPTGYTA 540
541 EQQSRASYDA HKTGHTGKDY DEHFLSVTRL DNVLELYTHF DDTEVLPHTS VLKFLTTLTM 600
601 FDIDLFNELN ATSFKYIPDC TMHRPKERTS SFNNTAHETG SEKTSGIKHI TQGLKKLTSL 660
661 PSSTKKTVKF VKMLLRNLNG NQAVSDVALL DTMRALLSFF TMTSAVFLVD RNLPSVLFAK 720
721 RLIPIMGTNL SVGQDWNSKI NNSLMVCLKK NSTTFVQLQL IFFSSAIQFD HELLLARLSI 780
781 DTMANNLNMQ KLCLYTEGFR IFFDIPSKKE LRKAIAVKIS KFFKTLFSII ADILLQEFPY 840
841 FDEQITDIVA SILDGTIINE YGTKKHFKGS SPSLCSTTRS RSGSTSQSSM TPVSPLGLDT 900
901 DICPMNTLSL VGSSTSRNSD NVNSLNSSPK NLSSDPYLSH LVAPRARHAL GGPSSIIRNK 960
961 IPTTLTSPPG TEKSSPVQRP QTESISATPM AITNSTPLSS AAFGIRSPLQ KIRTRRYSDE1020
1021 SLGKFMKSTN NYIQEHLIPK DLNEATLQDA RRIMINIFSI FKRPNSYFII PHNINSNLQW1080
1081 VSQDFRNIMK PIFVAIVSPD VDLQNTAQSF MDTLLSNVIT YGESDENISI EGYHLLCSYT1140
1141 VTLFAMGLFD LKINNEKRQI LLDITVKFMK VRSHLAGIAE ASHHMEYISD SEKLTFPLIM1200
1201 GTVGRALFVS LYSSQQKIEK TLKIAYTEYL SAINFHERNI DDADKTWVHN IEFVEAMCHD1260
1261 NYTTSGSIAF QRRTRNNILR FATIPNAILL DSMRMIYKKW HTYTHSKSLE KQERNDFRNF1320
1321 AGILASLSGI LFINKKILQE MYPYLLDTVS ELKKNIDSFI SKQCQWLNYP DLLTRENSRD1380
1381 ILSVELHPLS FNLLFNNLRL KLKELACSDL SIPENESSYV LLEQIIKMLR TILGRDDDNY1440
1441 VMMLFSTEIV DLIDLLTDEI KKIPAYCPKY LKAIIQMTKM FSALQHSEVN LGVKNHFHVK1500
1501 NKWLRQITDW FQVSIAREYD FENLSKPLKE MDLVKRDMDI LYIDTAIEAS TAIAYLTRHT1560
1561 FLEIPPAASD PELSRSRSVI FGFYFNILMK GLEKSSDRDN YPVFLRHKMS VLNDNVILSL1620
1621 TNLSNTNVDA SLQFTLPMGY SGNRNIRNAF LEVFINIVTN YRTYTAKTDL GKLEAADKFL1680
1681 RYTIEHPQLS SFGAAVCPAS DIDAYAAGLI NAFETRNATH IVVAQLIKNE IEKSSRPTDI1740
1741 LRRNSCATRS LSMLARSKGN EYLIRTLQPL LKKIIQNRDF FEIEKLKPED SDAERQIELF1800
1801 VKYMNELLES ISNSVSYFPP PLFYICQNIY KVACEKFPDH AIIAAGSFVF LRFFCPALVS1860
1861 PDSENIIDIS HLSEKRTFIS LAKVIQNIAN GSENFSRWPA LCSQKDFLKE CSDRIFRFLA1920
1921 ELCRTDRTID IQVRTDPTPI AFDYQFLHSF VYLYGLEVRR NVLNEAKHDD GDIDGDDFYK1980
1981 TTFLLIDDVL GQLGQPKMEF SNEIPIYIRE HMDDYPELYE FMNRHAFRNI ETSTAYSPSV2040
2041 HESTSSEGIP IITLTMSNFS DRHVDIDTVA YKFLQIYARI WTTKHCLIID CTEFDEGGLD2100
2101 MRKFISLVMG LLPEVAPKNC IGCYYFNVNE TFMDNYGKCL DKDNVYVSSK IPHYFINSNS2160
2161 DEGLMKSVGI TGQGLKVLQD IRVSLHDITL YDEKRNRFTP VSLKIGDIYF QVLHETPRQY2220
2221 KIRDMGTLFD VKFNDVYEIS RIFEVHVSSI TGVAAEFTVT FQDERRLIFS SPKYLEIVKM2280
2281 FYYAQIRLES EYEMDNNSST SSPNSNNKDK QQKERTKLLC HLLLVSLIGL FDESKKMKNS2340
2341 SYNLIAATEA SFGLNFGSHF HRSPEVYVPE DTTTFLGVIG KSLAESNPEL TAYMFIYVLE2400
2401 ALKNNVIPHV YIPHTICGLS YWIPNLYQHV YLADDEEGPE NISHIFRILI RLSVRETDFK2460
2461 AVYMQYVWLL LLDDGRLTDI IVDEVINHAL ERDSENRDWK KTISLLTVLP TTEVANNIIQ2520
2521 KILAKIRSFL PSLKLEAMTQ SWSELTILVK ISIHVFFETS LLVQMYLPEI LFIVSLLIDV2580
2581 GPRELRSSLH QLLMNVCHSL AINSALPQDH RNNLDEISDI FAHQKVKFMF GFSEDKGRIL2640
2641 QIFSASSFAS KFNILDFFIN NILLLMEYSS TYEANVWKTR YKKYVLESVF TSNSFLSARS2700
2701 IMIVGIMGKS YITEGLCKAM LIETMKVIAE PKITDEHLFL AISHIFTYSK IVEGLDPNLD2760
2761 LMKHLFWFST LFLESRHPII FEGALLFVSN CIRRLYMAQF ENESETSLIS TLLKGRKFAH2820
2821 TFLSKIENLS GIVWNEDNFT HILIFIINKG LSNPFIKSTA FDFLKMMFRN SYFEHQINQK2880
2881 SDHYLCYMFL LYFVLNCNQF EELLGDVDFE GEMVNIENKN TIPKILLEWL SSDNENANIT2940
2941 LYQGAILFKC SVTDEPSRFR FALIIRHLLT KKPICALRFY SVIRNEIRKI SAFEQNSDCV3000
3001 PLAFDILNLL VTHSESNSLE KLHEESIERL TKRGLSIVTS SGIFAKNSDM MIPLDVKPED3060
3061 IYERKRIMTM ILSRMSCSA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
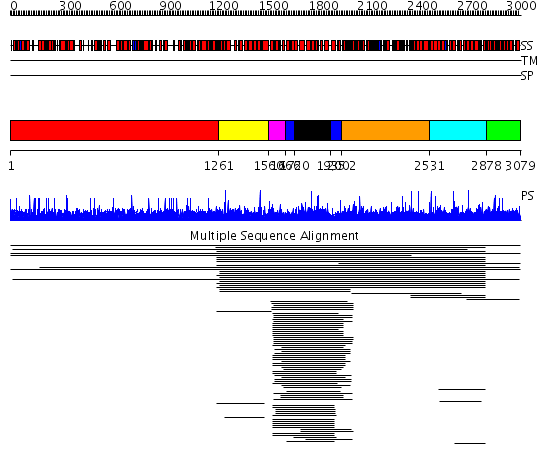
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1260] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [1261..1559] | 2.015945 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1560..1665] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1666..1719] [1935..2001] |
312.457575 | GAP related domain of neurofibromin | |
| 5 | View Details | [1720..1934] | 312.457575 | GAP related domain of neurofibromin | |
| 6 | View Details | [2002..2530] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [2531..2877] | 2.014907 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [2878..3079] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)