













| General Information: |
|
| Name(s) found: |
FAB1 /
YFR019W
[SGD]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2278 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA late endosome [IDA vacuolar membrane [IDA vacuole [IDA |
| Biological Process: |
vacuole organization
[IMP late endosome to vacuole transport [IMP phospholipid metabolic process [IGI negative regulation of transcription from RNA polymerase II promoter by glucose [IMP Golgi to vacuole transport [IMP |
| Molecular Function: |
1-phosphatidylinositol-3-phosphate 5-kinase activity
[IDA phosphatidylinositol-3-phosphate binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSEEPHASI SFPDGSHVRS SSTGTSSVNT IDATLSRPNY IKKPSLHIMS TSTTSTTTDL 60
61 VTNPILSNIS VPKISPPTSS SIATATSTSH VTGTASHSNI KANANTSTSV NKKNLPPTTS 120
121 GRIPSSTIKR YPSRYKPSHS LQLPIKNDSN FKRSSIYASK STVTAIPIRN NRPISMQNSY 180
181 ARTPDSDHDD VGDEVSSIKS ASSSLTASLS KSFLFAFYNN RKKDKTSNNG VLSKEYWMKD 240
241 ESSKECFSCG KTFNTFRRKH HCRICGQIFC SSCTLLIDGD RFGCHAKMRV CYNCYEHADT 300
301 YEDSSDEEND STMQLNEPRS RSRSRSSNTN PYSHSHSHLH LISQDNHNGT DLHDPVAATD 360
361 NPQQQNEVYL LNDDDVQSIM TSGEDSKLFI STPPPPPKMA IPATKQGGSL EISFDSENDR 420
421 ALHYQDDNPG RHHHLDSVPT RYTIRDMDNI SHYDTNSNST LRPHYNTNNS TITINNLNNT 480
481 TSNNSNYNNT NSNSNINNPA HSLRRSIFHY VSSNSVNKDS NNSSATPASS AQSSSILDPA 540
541 NRIIGNYAHR NYKFKFNYNS KGPSQQNDTA NGNNDNNNNN NNNNNNNNNN SASGIADNNN 600
601 IPSNDNGTTF TLDKKKRNPL TKSKSTSAYL EYPLNEEDSS EDEGSMSIYS VLNDDHKTDN 660
661 PIRSMRNSTK SFQRAQASLQ RMRFRRKSKS KHFPNNSKSS IYRDLNFLTN STPNLLSVVS 720
721 DDNLYDDSSP LQDKASSSAA SRLTDRKFSN SSGSNNNSNS NSNINTDPWK RIASISGFKL 780
781 KKEKKRELNE VSLLHMHALL KQLLNDQEIS NLQEWITLLD GALRKVLRTI LNARDLNTLD 840
841 FRQTYVKIKR ISGGSPQNSE YIDGVVFSKA LPSKTMPRHL KNPRILLIMF PLEYQKNNNH 900
901 FLSIESVFRQ EREYLDKLVS RLKSLHPDII YVGANVSGYA LELLNDSGIV VQFNMKPQVI 960
961 ERIAKLTEAD IAISVDKLAT NIKMGECETF EVKSYIYGNI SKTYTFLRGC NPELGGTILL1020
1021 RGDSLENLRK IKQVSEFMVY AIFSLKLESS FFNDNFIQLS TDVYLKRAES KKLQVFEGYF1080
1081 ADFLIKFNNR ILTVSPTVDF PIPFLLEKAR GLEKKLIERI NQYESESDLD RQTQLNMLQG1140
1141 LESTITKKHL GNLIKFLHEM EIENLELEFQ KRSRQWEVSY SSSQNLLGTG SHQSITVLYS1200
1201 MVSTKTATPC VGPQIVTIDY FWDSDISIGQ FIENVVGTAR YPCQQGCNGL YLDHYRSYVH1260
1261 GSGKVDVLIE KFQTRLPKLK DIILTWSYCK KCGTSTPILQ ISEKTWNHSF GKYLEVMFWS1320
1321 YKDSVTGIGK CPHDFTKDHV KYFGYNDLVV RLEYSDLEVH ELITPPRKIK WKPHIDIKLK1380
1381 VELYYKILEK INNFYGSVLS RLERIKLDSM TKDKVLSGQA KIIELKSNAT EEQKLMLQDL1440
1441 DTFYADSPCD QHLPLNLVIK SLYDKAVNWN STFAIFAKSY LPSETDISRI TAKQLKKLFY1500
1501 DSSRKDSEDK KSLHDEKAKT RKPEKNELPL EGLKDVEKPK IDSKNTTENR DRTNEPQNAV1560
1561 TITTFKDDTP IIPTSGTSHL TVTPSASSVS SSLTPQTEER PPISRSGTGI SMTHDKSTRP1620
1621 NIRKMSSDSS LCGLASLANE YSKNNKVSKL ATFFDQMHFD ALSKEFELER ERERLQLNKD1680
1681 KYQAIRLQTS TPIVEIYKNV KDAVDEPLHS RSSGNNLSSA NVKTLEAPVG EHSRANNCNP1740
1741 PNLDQNLETE LENSISQWGE NILNPSGKTT ASTHLNSKPV VKETSENPKS IVRESDNSKS1800
1801 EPLPPVITTT TVNKVESTPQ PEKSLLMKTL SNFWADRSAY LWKPLVYPTC PSEHIFTDSD1860
1861 VIIREDEPSS LIAFCLSTSD YRNKMMNLNV QQQQQQQTAE AAPAKTGGNS GGTTQTGDPS1920
1921 VNISPSVSTT SHNKGRDSEI SSLVTTKEGL LNTPPIEGAR DRTPQESQTH SQANLDTLQE1980
1981 LEKIMTKKTA THLRYQFEEG LTVMSCKIFF TEHFDVFRKI CDCQENFIQS LSRCVKWDSN2040
2041 GGKSGSGFLK TLDDRFIIKE LSHAELEAFI KFAPSYFEYM AQAMFHDLPT TLAKVFGFYQ2100
2101 IQVKSSISSS KSYKMDVIIM ENLFYEKKTT RIFDLKGSMR NRHVEQTGKA NEVLLDENMV2160
2161 EYIYESPIHV REYDKKLLRA SVWNDTLFLA KMNVMDYSLV IGIDNEGYTL TVGIIDFIRT2220
2221 FTWDKKLESW VKEKGLVGGA SVIKQPTVVT PRQYKKRFRE AMERYILMVP DPWYWEGN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
AEP3 CDC20 FAB1 FOL2 GZF3 LSP1 NOP13 PIL1 PKH1 PRP16 PSK2 RGC1 RPL9B YLR179C YMR086W YTA6 |
| View Details | Gavin AC, et al. (2002) |
|
ACC1 ADA2 ADH1 AVT4 BEM2 CCT2 CDC25 COG3 CSE2 ENO2 FAB1 FBA1 GAL11 GCN5 HRK1 MDS3 MED1 MED11 MED2 MED4 MED6 MED7 MED8 MIA40 MLP1 MRPL3 MRPL6 NUT1 NUT2 PGD1 PGK1 RGR1 ROX3 RPG1 RPO21 SAP155 SAP185 SAP190 SIN4 SIT4 SPT7 SRB2 SRB4 SRB5 SRB6 SRB7 SRB8 SRP1 SSE2 SSN2 SSN3 SSN8 SWT21 TAF6 TIF1, TIF2 YIL077C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
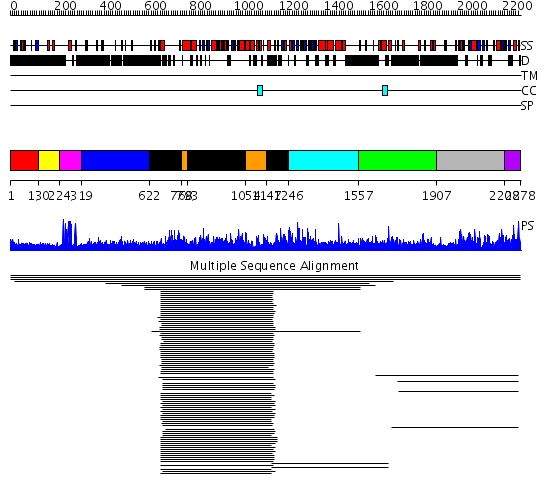
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 3.036995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [130..223] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [224..318] | 93.014858 | Eea1 | |
| 4 | View Details | [319..621] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [622..767] [793..1053] [1147..1245] |
266.9794 | Thermosome, E domain; Thermosome, A-domain; Thermosome, I domain | |
| 6 | View Details | [768..792] [1054..1146] |
266.9794 | Thermosome, E domain; Thermosome, A-domain; Thermosome, I domain | |
| 7 | View Details | [1246..1556] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1557..1906] | 1.023926 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1907..2207] | 180.325697 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta | |
| 10 | View Details | [2208..2278] | 9.06 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta | |
| 11 | View Details | [1834..1938] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1939..2207] | 87.154902 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta | |
| 13 | View Details | [2208..2278] | 3.05 | No description for 2gk9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)