













| General Information: |
|
| Name(s) found: |
AEP3 /
YPL005W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 606 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA extrinsic to membrane [IDA mitochondrial inner membrane [IDA |
| Biological Process: |
mRNA metabolic process
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTLRCLTQA LSKSGREAPK LYQKVIFPGL FREGIPIANV KKVDEKIIDS PTSTSVNGEA 60
61 KKIVRHGVKY EREQVKEYLS SLPTLTLSRK QIRDDYDEER AKRMYMFSKQ TNSSNKFQKL 120
121 LTAKSQEFTR ELLTLLIDCT SNEKNSGPER FTRKFLKFSN DEIPPLPDFS KNPQLFENYI 180
181 GILSHTKFNF RSSSKLNGIV RKMLRHLLHP TNKTTLPLRS AQVYNDSIYF FSEHFDFASC 240
241 REIFAQMKAE GTKPNTITFN LLLRNVVKNS HIRKTKHPDD EVLFYLRSMR NHGVFADVIT 300
301 WTTCYNFLRD EVSRQLYIVQ MGEHLGNFNV NFVYTVLRNG DYRAEDCLKV LAANSLPISR 360
361 KTFYLCIERL LNEEQLETAS KLLDYGFQHL KSNFKLDSEA MNHFMRVFAN KGRSDLAFLC 420
421 YNTCRKIYKI KPDSQTFEML FKALVRNGNT KNFGAVLQYI KDLKVSEGFG LRTSYWRTKA 480
481 DSIFKFGSPN TLSEKSIEKA RKLLGNLIAS EGEFSWKIWK ESDSSQKKIL RFLGCIPTTL 540
541 RCTNTAQDHQ KPTNLPSNIS QKKREYRNRV KAIATKAALE KRMAYIKDND VAFKKELVKR 600
601 RIVGEV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
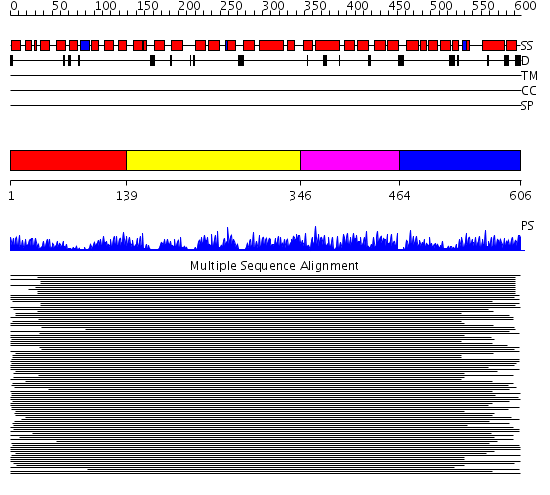
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 2.700995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [139..345] | 8.802996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [346..463] | 3.522879 | Designed protein cTPR3 | |
| 4 | View Details | [464..606] | 2.748986 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)